2019 Volume 44 Issue 5 Pages 327-333
2019 Volume 44 Issue 5 Pages 327-333
Metallothionein (MT) is a low-molecular-weight, cysteine-rich, and metal-binding protein that protects cells from the cytotoxic effects of heavy metals and reactive oxygen species. Previously, we found that transcriptional induction of endothelial MT-1A was mediated by not only the metal-regulatory transcription factor 1 (MTF-1)-metal responsive element (MRE) pathway but also the nuclear factor-erythroid 2-related factor 2 (Nrf2)-antioxidant response element/electrophile responsive element (ARE) pathway, whereas that of MT-2A was mediated only by the MTF-1-MRE pathway, using the organopnictogen compounds tris(pentafluorophenyl)stibane, tris(pentafluorophenyl)arsane, and tris(pentafluorophenyl)phosphane as molecular probes in vascular endothelial cells. In the present study, we investigated the binding sites of MTF-1 and Nrf2 in the promoter regions of MTs in cultured bovine aortic endothelial cells treated with these organopnictogen compounds. We propose potential mechanisms underlying transcriptional induction of endothelial MT isoforms. Specifically, both MRE activation by MTF-1 and that of ARE in the promoter region of the MT-2A gene by Nrf2 are involved in transcriptional induction of MT-1A, whereas only MRE activation by MTF-1 or other transcriptional factor(s) is required for transcriptional induction of MT-2A in vascular endothelial cells.
Metallothionein (MT) is a low-molecular-weight, cysteine-rich, and metal-binding protein (Kägi and Vallee, 1960). MT is induced by heavy metals and protects cells against the cytotoxicity of heavy metals and reactive oxygen species and is involved in zinc homeostasis (Davis and Cousins, 2000). Additionally, MT has anti-inflammatory effects and thus is considered a cytoprotective protein. Although inorganic zinc is a typical inducer of MT, we demonstrated that inorganic zinc cannot induce MT in vascular endothelial cells, which cover the luminal surface of blood vessels (Kaji et al., 1992; Fujie et al., 2016a).
It is thought that the metal response element (MRE)-binding transcription factor-1 (MTF-1) is crucially involved in MT induction. The transcriptional factor is activated when zinc ion binds to the zinc finger domains of the molecule (Bittel et al., 1998) and activated MTF-1 binds to the MRE, the consensus sequences of which are in MT gene promoter regions (Heuchel et al., 1994). In contrast, the antioxidant response element (ARE) contains consensus sequences and is activated by the transcription factor nuclear factor-erythroid 2-related factor 2 (Nrf2) (Itoh et al., 1997; Ohtsuji et al., 2008) in the MT gene promoter regions, although its role in MT isoform induction remains unclear.
We found that Nrf2 contributes in part to inducing MT by cadmium in cultured bovine aortic endothelial cells (Shinkai et al., 2016). However, because cadmium induces the expression of both isoforms of MT, MT-1 and MT-2, it is difficult to analyze the detailed mechanisms. Recently, in a study of bioorganometallics (Fujie et al., 2016b), we found that tris(pentafluorophenyl)stibane (Sb35), tris(pentafluorophenyl)arsane (As35), and tris(pentafluorophenyl)phosphane (P35) cause transcriptional induction of endothelial MT (Fujie et al., 2016c).
Specifically, both Sb35 and As35 induce the expression of MT-1A and MT-2A, whereas P35 increases the expression of only MT-2A in cells. Transcriptional induction of endothelial MT-1A was mediated by not only the MTF-1- MRE pathway but also the Nrf2- ARE pathway, whereas that of MT-2A was mediated only by the MTF-1-MRE pathway (Fujie et al., 2016c, 2016d). In the present study, we investigated the binding sites of MTF-1 and Nrf2 activated by Sb35, As35, and P35 in the promoter regions of MTs in cultured vascular endothelial cells.
Bovine aortic endothelial cells were purchased from Cell Applications (San Diego, CA, USA). The following materials were also used: Dulbecco’s modified Eagle’s medium (DMEM) and calcium- and magnesium-free phosphate buffered saline (CMF-PBS) from Nissui Pharmaceutical (Tokyo, Japan); fetal bovine serum, Halt™ Protease Inhibitor Cocktail (100×), and Sheared Salmon Sperm DNA from Thermo Fisher Scientific (Waltham, MA, USA); Protein G beads from Tamagawa Seki (Nagano, Japan); and GeneAce SYBR qPCR Mixα from Nippon Gene (Tokyo, Japan). Other reagents were purchased from Nacalai Tesque (Kyoto, Japan).
Synthesis of Sb35, As35, and P35Sb35, As35, and P35 were synthesized as described previously (Fild et al., 1964; Kant et al., 2008; Schäfer et al., 2011; Jiang et al., 2013).
Cell culture and treatmentBovine aortic endothelial cells were cultured at 37°C in 5% CO2 in DMEM supplemented with 10% fetal bovine serum until confluence. The medium was removed, and the cells were washed twice with serum-free DMEM; the cells were exposed to Sb35 (50, 100, and 200 μM), As35 (5, 10, and 20 μM), or P35 (5, 10, and 20 μM) in serum-free DMEM for 3 hr at 37°C. Because the organopnictogen compounds used in this study were insoluble in water, they were dissolved in dimethyl sulfoxide and then added to the culture medium; the final concentration of dimethyl sulfoxide was less than 0.5%.
Chromatin immunoprecipitation (ChIP) assayBovine aortic endothelial cells were treated with Sb35, As35, or P35 for 3 hr. The DNA-proteins were cross-linked by incubating the cells with 1% formaldehyde at 37°C for 10 min. Excess formaldehyde was quenched with 0.125 M glycine at 37°C for 5 min. After washing with ice-cold CMF-PBS, the cells were scraped in ice-cold CMF-PBS and collected by centrifugation. The pellet was lysed in sodium dodecyl sulfate (SDS)-lysis buffer (50 mM Tris-HCl, 10 mM EDTA, 1% SDS, pH 8.1) supplemented with protease inhibitors and sonicated for 75 cycles (3-s pulse and 1-s rest, on ice) using an Ultrasonic Homogenizer (Microtec, Chiba, Japan). Sheared chromatin was diluted in ChIP dilution buffer (25 mM Tris-HCl, 150 mM sodium chloride, 1% Triton X-100, 0.1% sodium deoxycholate, 2 mM EDTA, pH 8.1) containing salmon sperm DNA (0.5 mg/mL) and a portion of the chromatin solution was stored as input DNA. The remaining chromatin was immunoprecipitated using protein G beads coated with normal rabbit IgG (#2729; Cell Signaling, Danvers, MA, USA), Nrf2 (NBP1-32822; Novus Biologicals, Littleton, CO, USA), or MTF-1 (NBP1-86380; Novus Biologicals) antibodies for 6 hr at 4°C. After washing, protein-DNA complexes were eluted from the antibody with elution buffer (50 mM Tris-HCl, 10 mM EDTA, 1% SDS, pH 8.1) and reverse cross-linked by heating overnight at 65°C. The DNA samples were purified using proteinase K treatment followed by phenol-extraction and ethanol precipitation, and then analyzed by real-time PCR using GeneAce SYBR qPCR Mixα on a StepOnePlus RT-PCR system (Thermo Fisher Scientific). The thermal cycling parameters were 50°C for 2 min, 95°C for 10 min, and 50 cycles of 95°C for 15 sec and 60°C for 1 min. The primers for ChIP analysis are listed in Table 1. Relative DNA levels were calculated from a standard curve using serial dilutions of input DNA and normalized with input DNA from a corresponding sample.
| Target region | Forward primer (5′→ 3′) | Reverse primer (5′→ 3′) | |
|---|---|---|---|
| MT-1A | MRE | ACAAAGTTCCTCCGTGTTGG | GTCGGATGGGAGGCGTGCTG |
| MT-1E | MRE | AGACTCTTGCGCTGGGCTC | CTCGCTCGCTGGGTTTGTAA |
| MT-2A | MRE | ACGTAGGGCGGCTTCTGGGA | GCAAAGGATGGCGAGGGTGTC |
| GCLM | ARE | GGGACGGGTAACGGTTAGCAA | CGGTAGAGTGGACTCCCAACTGA |
| MT-1A | ARE1&2 | TCTCACCTCCTGGCCACACA | AGCACTGAGCTGGCAGTCAGAA |
| ARE3 | GGCTTGCCCAGGGTCAC | ACAATCACTTTGCTGCTTTCG | |
| MT-1E | ARE1 | ATGATGCTGCCACACCATTTG | GGCGAGTAGGTGAGACAGCTATAGA |
| ARE2&3 | CACTTCCTGCCACATATCCTGA | CCTGGTTAGGCCACAGTCCA | |
| MT-2A | ARE1 | CGTGTGCACAGCTCGGTGA | GGAGCTGGGACGAGTGCAAA |
| ARE2 | CCATCCTAGCCACGACTCTGGTA | CCAACAGATGCTGAAGTCCCTCTA | |
| ARE3&4 | CACAGGAAGCACCAGGAAGGAA | TCTTCACAGTTCAGCTCACACATCA | |
| ARE5 | TAAGGATACTGTCACAGGGGAAAGA | GTGTGAGTCATCAGTGTGAGGCAA | |
A map of the MRE and ARE regions in the bovine MT isoforms and sequences of MRE and ARE in the promoter of the bovine MT-1A, MT-1E, and MT-2A genes are shown in Fig. 1. We tested bovine MT-1A, MT-1E, and MT-2A promoter regions containing six, five, and six, respectively, MRE sequences and three, three, and five, respectively, ARE sequences.
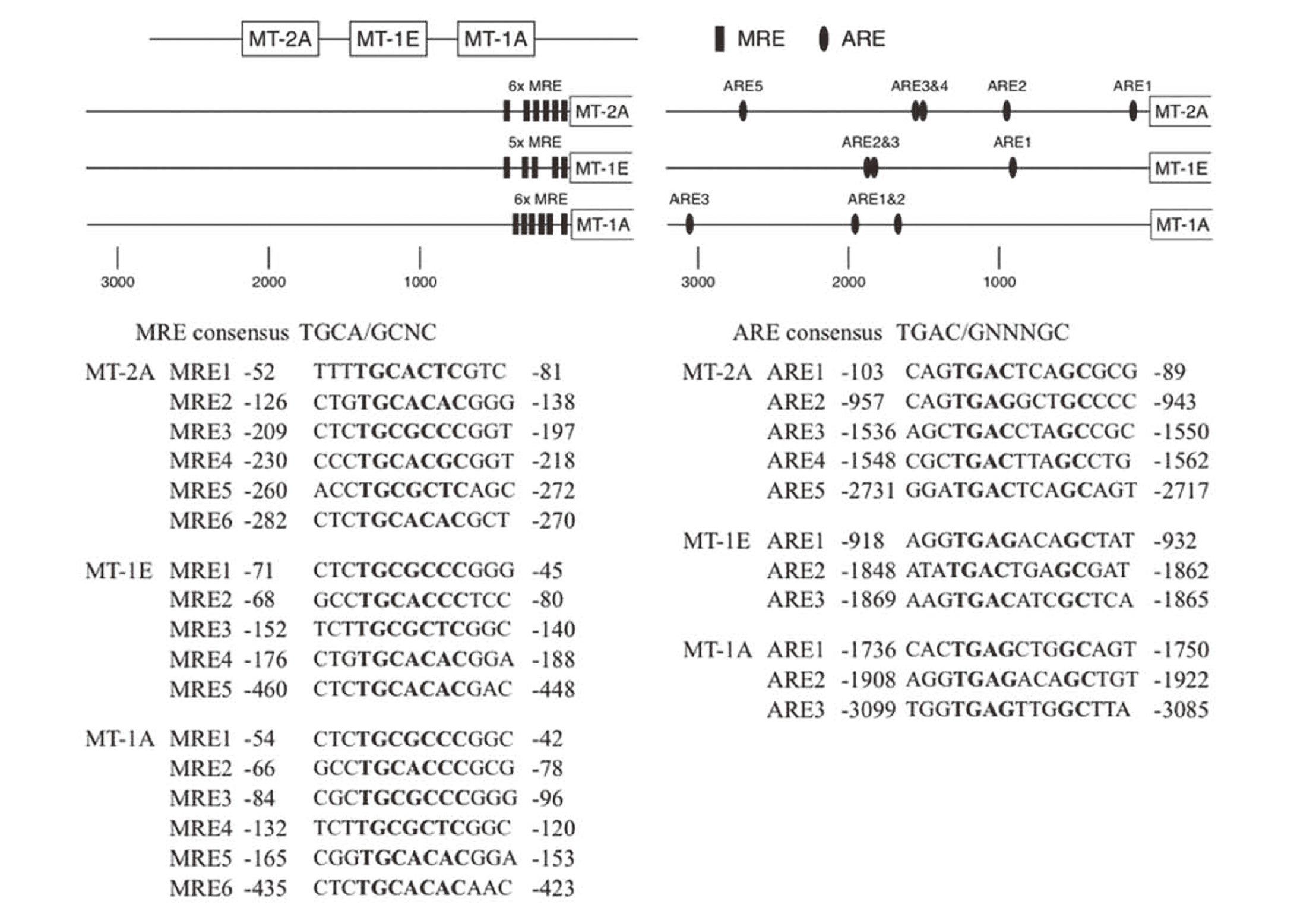
Map of MRE and ARE regions in bovine MT promoter and corresponding MRE and ARE sequences.
The molecular structures of Sb35, As35, and P35 are shown in Fig. 2A. We found that MTF-1 was recruited to the MREs of the promoter regions of MT-1A, MT-1E, and MT-2A by Sb35 and As35 (Figs. 2B and 2C, respectively), whereas P35 failed to recruit MTF-1 to the MREs (Fig. 2D). Previously, we reported that P35 as well as Sb35 and As35 induce the expression of endothelial MT-2A by activating MRE (Fujie et al., 2016c). Therefore, our present and previous data suggest that Sb35 and As35 activate MTF-1, leading to the activation of MREs in the promoter region of MT, whereas P35 also activates the MREs by transcriptional factor(s) other than MTF-1 such as hypoxia inducible factor-1α (Dubé et al., 2011). As a result, these organopnictogen compounds induce the expression of MT-2A in vascular endothelial cells. Activation of MREs specifically by MTF-1 may be required for the expression of endothelial MT-1A, although P35 may promote the recruitment of MTF-1 to other MREs other than the MT promoter regions tested in this study.
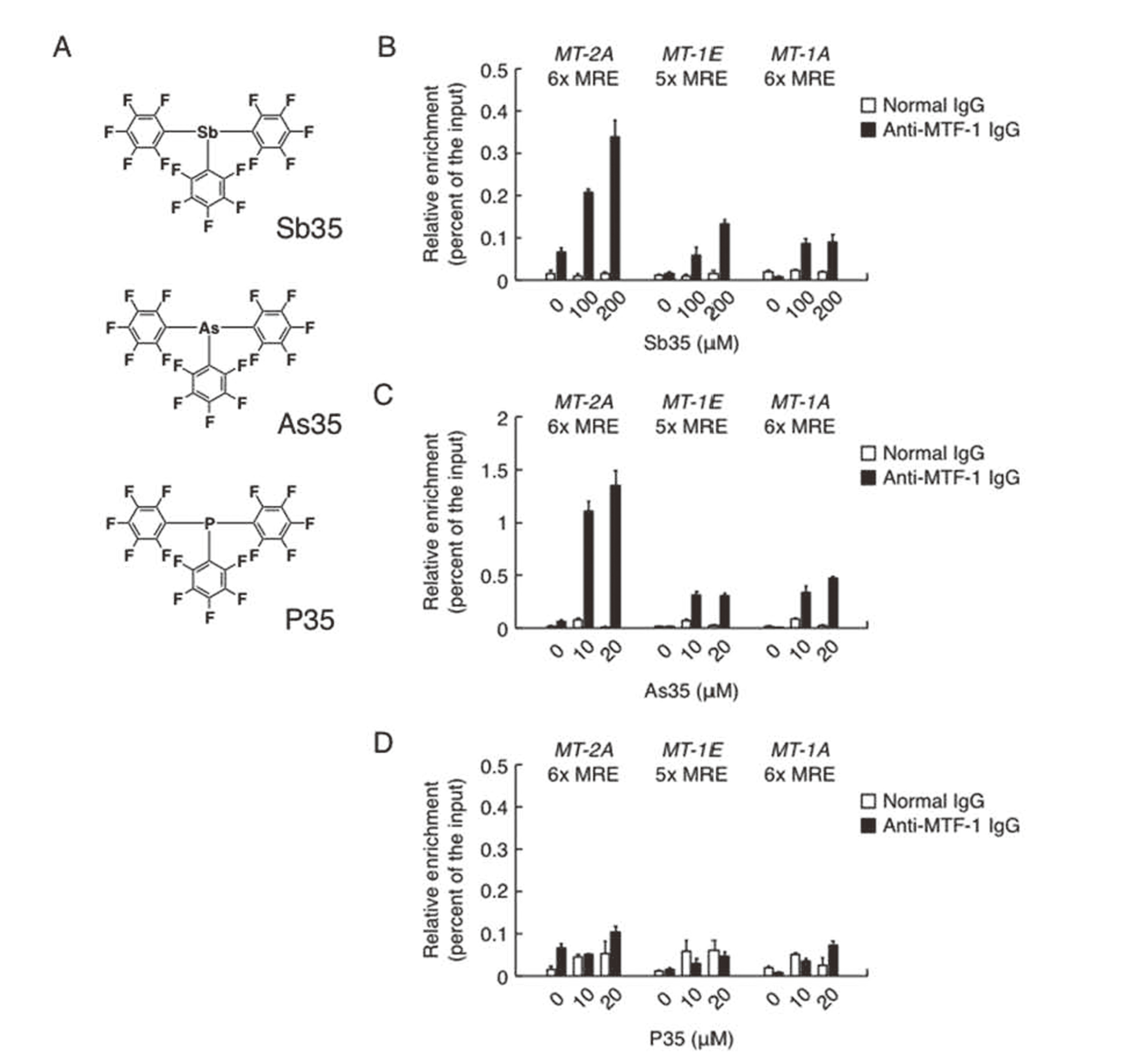
Recruitment of MTF-1 to the promoter regions of MT genes in vascular endothelial cells. [A] Structures of organopnictogen compounds used in this study. Sb35, tris(pentafluorophenyl)stabane; As35, tris(pentafluorophenyl)arsane; P35, tris(pentafluorophenyl)phosphane. Bovine aortic endothelial cells were treated with or without [B] Sb35 (100 and 200 µM), [C] As35 (10 and 20 µM), or [D] P35 (10 and 20 µM) for 3 hr. Enrichment of the promoter DNA precipitated was determined by the ChIP assay with the indicated antibodies. Values represents the means ± SE of two independent experiments.
Figure 3 shows the recruitment of Nrf2 to AREs in the promoter regions of MT genes in vascular endothelial cells. Treatment with Sb35, As35, and P35 significantly increased the recruitment of Nrf2 to AREs in the promoter region of GCLM, a target gene of Nrf2 in the cells (Fig. 3A), suggesting that these organopnictogen compounds activated Nrf2 as reported previously (Fujie et al., 2016c). It was demonstrated that Sb35 (Fig. 3B), As35 (Fig. 3C), and P35 (Fig. 3D) recruited Nrf2 to MT-2A ARE5 but not the other AREs of MT-1A and MT-1E in the MT promoter regions. Because transcriptional induction of endothelial MT-1A by Sb35 was mediated by the Nrf2-ARE pathway and MTF-1-MRE pathway (Fujie et al., 2016c), the involvement of the Nrf2-ARE pathway in inducing MT-1A may be mediated by the recruitment of Nrf2 to MT-2A ARE5 in vascular endothelial cells. P35, which activates MRE without recruiting MTF-1 to MREs and induces only MT-2A expression, also activates Nrf2 and promotes the recruitment of Nrf2 to MT-2A ARE5, supporting the hypothesis that endothelial MT-1A expression requires the activation of MREs specifically by MTF-1 but not the other transcriptional factors.

Recruitment of Nrf2 to the promoter regions of MT genes in vascular endothelial cells. Bovine aortic endothelial cells were treated with or without [A] Sb35 (100 and 200 μM), As35 (10 and 20 μM), or P35 (10 and 20 μM), [B] Sb35 (100 and 200 µM), [C] As35 (10 and 20 µM), or [D] P35 (10 and 20 µM) for 3 hr. Enrichment of the promoter DNA precipitated was determined by the ChIP assay with the indicated antibodies. Values represents the means ± SE of two independent experiments.
In conclusion, based on our previous and present data, we predicted the mechanisms underlying transcriptional induction of endothelial MT isoforms. Specifically, both MRE activation by MTF-1 and that of ARE in the promoter region of the MT-2A gene by Nrf2 are involved in the transcriptional induction of MT-1A, whereas only MRE activation by MTF-1 or other transcriptional factor(s) is required for transcriptional induction of MT-2A in vascular endothelial cells. It is possible that involvement of Nrf2 in inducing endothelial MT-1A is attributed to epigenetic regulation such as changes the chromatin structure around MTs, as it was reported that Nrf2 activation caused chromatin remodeling around the human heme oxygenase-1 promoter by Brahma-related gene 1 (Zhang et al., 2006; Maruyama et al., 2013). Although further studies are required to clarify the detailed mechanisms of endothelial MT isoform induction, our previous and present studies clearly showed that bioorganometallics, which can be used to analyze biological systems using organic-inorganic hybrid molecules (Fujie et al., 2016b), is useful for analyzing the mechanisms underlying MT induction in vascular endothelial cells.
This work was supported by a Grant-in-Aid for Challenging Exploratory Research JP15K14992 (to T. K.) and Grant-in-Aid for Scientific Research (C) JP18K06638 (to C. Y.) from the Japan Society for the Promotion of Science.
Conflict of interestThe authors declare that there is no conflict of interest.