2020 Volume 45 Issue 12 Pages 751-761
2020 Volume 45 Issue 12 Pages 751-761
The phorbol 12-myristate 13-acetate (PMA)-induced U937 cell line has been widely used as an in vitro model for studying the functions of human macrophages. However, there are several concentrations of PMA commonly used to drive the differentiation of monocytic cell line to macrophage. Also, the expression of microRNA-155 (miR-155) and miR-125b in PMA-treated human monocytic cell line has not yet been reported. The five usual concentrations of PMA for stimulating macrophage differentiation are 10, 25, 50, 100, and 200 nM. In this study we compared the expression levels of miR-155, miR-125b and their related genes involved in macrophage functions in U937-derived cells after treatment with those five concentrations. The morphological study results showed that the five concentrations of PMA could induce macrophage differentiation in a similar manner. Moreover, cell proliferation and viability were not significantly different among these five conditions excepted the lower cell viability at 200 nM of PMA treatment. The five concentrations of PMA could upregulate the expression of miR-155 and miR-125b and increase the phagocytic activity of U937-derived cells in dose-reversal manner. The upregulation of miR-155 was correlated with increased expression levels of TNFα and decreased expression levels of BACH1 and CEBPβ, while the reduction of IRF4 was correlated with increased expression levels of miR-125b. Our study found that PMA could stimulate macrophage differentiation in a broad range of concentrations, however, the lower concentration could upregulate the higher expression of both miR-155 and miR-125b, and that correlated with the phagocytic functional activity of U937-derived macrophages.
Macrophages play a pivotal role in the immune system, mainly in the clearance of various types of pathogens within the human body via phagocytosis (Gordon and Martinez-Pomares, 2017). Macrophages can be activated into the classical pro-inflammatory (M1) or alternatively regenerative (M2) macrophages, depending on environmental factors. In general, macrophages can be activated to the M1 state by interferon gamma (IFN-γ), granulocyte macrophage-colony stimulating factor (GM-CSF) and lipopolysaccharide (LPS) and to the M2 state by macrophage-colony stimulating factor (M-CSF), interleukin-4 (IL-4), IL-10, IL-13, transforming growth factor beta (TGF-β), and glucocorticoids (Martinez et al., 2008; Vogel et al., 2014). Recent studies have found that miRs played important roles as stimulators of macrophage activation. One study found that miR-155 was upregulated in activated monocytes or macrophages induced by inflammatory stimulators such as LPS and IFN-γ (Jablonski et al., 2016). Another study reported that miR-155 also had a potential role in inducing TNF-α production in LPS-stimulated macrophages via increased mRNA stability (Bala et al., 2011). Another study found it could enhance phagocytic activity of β-thalassemia/HbE monocytes via targeting of BACH1 (Srinoun et al., 2017). The target genes of miR-155 which are involved in immunologic cell-suppressing gene expression include the CCAAT/enhancer-binding protein beta (CEBPβ) (He et al., 2009), inositol polyphosphate-5-phosphatase D (INPP5D) or Src homology-2 domain-containing inositol 5-phosphatase 1 (SHIP1) (O’Connell et al., 2009), Jumonji and AT-Rich Interaction Domain containing 2 (JARID2) (Norfo et al., 2014) and BACH1 (Yin et al., 2008). A previous study found that CEBPβ translation was suppressed by miR-155 through interaction with the 3’UTR of CEBPβ mRNA and miR-155-regulated inflammatory cytokine production in tumor-associated macrophages via targeting CEBPβ (He et al., 2009). Another study found that miR-125b was enriched in macrophages and partly responsible for macrophage activation, at least partially by reducing IFN regulatory factor 4 (IRF4) levels (Chaudhuri et al., 2011). Our previous study found the higher miR-125b expression in CD14-positive monocytic cells of β- thalassemia patients and that correlated with the higher phagocytic activity (Kuno et al., 2019). IRF4 is a member of the IFN response factor family of transcription factors, which has a function to inhibit the inflammatory response (Honma et al., 2005; Negishi et al., 2005). Enhanced expression of miR-125b could drive macrophages to express an activated morphology, increase the expression of the co-stimulatory molecules CD40, CD86, and CD80, and elevate responsiveness to IFN-γ (Chaudhuri et al., 2011).
The U937 human monocytic cell line has been widely used as a study model to investigate human monocyte and macrophage biology and functions (Bradley et al., 2018). The differentiation of U937 monocytes into macrophage-like cells can be induced by PMA (Wu et al., 1994; Bradley et al., 2018). The mechanism of PMA to induce the differentiation of monocyte to macrophage involves NF-kB activation through the protein kinase C pathway (Krappmann et al., 2001). PMA also activates RhoA/ROCK signaling during monocyte to macrophage differentiation resulting in morphological changes and increased cell adherence (Yang et al., 2017). However, there is a wide range of PMA concentrations being used in the reported experiments, and this wide range could affect the macrophage status through mechanisms such as the increased transcription levels of adhesion molecules CD44, CD14, ICAM-1 or the extracellular matrix FN1 in high-dose PMA treatments (Song et al., 2015; Emirbayer et al., 2017). A previous study found that THP-1 macrophages differentiated in high concentrations of PMA rapidly died following infection whereas those differentiated in low concentrations of PMA survived and were able to control the intracellular Salmonella Typhimurium bacteria similar to primary human macrophages (Starr et al., 2018). Therefore, it is important to use the appropriate conditions to differentiate cell lines when using them as models for studying macrophage functions.
In this study, we examined the effects of five commonly-used concentrations of PMA on the expression of two related miRs (miR-155 and miR-125b) and their macrophage function-related genes including TNFα, BACH1, and CEBPβ which were regulated by miR-155 and IRF4 which was regulated by miR-125b. Moreover, the effect of PMA concentrations on the phagocytic function of macrophage was also determined.
U937 monocytic cells were cultured in RPMI1640 medium (GIBCO-Invitrogen, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS), L-glutamine, and antibiotic-antimycotic (GIBCO-Invitrogen) To promote the differentiation of the U937 cell line towards macrophage lineage, five concentrations of PMA (Sigma Aldrich, St. Louis, MO, USA) at 10, 25, 50, 100, and 200 nM resuspended in 500 µL of culture media were used to treat 3x105 cells of U937 in 24-well plates for 48 hr. PMA-untreated U937 cells were used as a control.
Cell proliferation and viabilityThe 3 x 105 cells of U937 were treated with 0, 10, 25, 50, 100, and 200 nM of PMA in 24-well plates for 24 and 48 hr. The effects of PMA concentrations on cell proliferation and viability were determined by trypan blue staining. Briefly, the cell suspensions were mixed with 0.4% trypan blue at a 10:1 ratio. Viable cells that could exclude the trypan blue appeared transparent while dead cells were stained blue. Cell numbers were counted on hemocytometer with a light microscope.
Cell differentiationThe effects of PMA concentrations on the differentiation of U937-derived cells were studied by determined cell morphology and the expression levels of the monocyte-macrophage surface antigen CD14. The 3 x 105 cells of U937 were treated with 0, 10, 25, 50, 100, and 200 nM of PMA for 48 hr in 24-well plates. The 1 x 105 of these cells were washed twice with phosphate-buffered saline (PBS) containing 2% FBS and then incubated with biotin-streptavidin PE antibody (eBioscience, San Diego, CA, USA) conjugated anti-CD14 at 1:300 dilution for 40 min. Cells were washed twice and then measured with flow cytometry (BD FACSCalibur™, BD Bioscience, San Jose, CA, USA). At least 10,000 cell signals were obtained in each sample and the data were analyzed by Cell Quest software (Becton Dickinson, San Jose, CA, USA). The cell morphology was studied using brightfield microscopy and May-Grünwald-Giemsa staining. The 5-10 x 104 of U937-treated cells were added to a cytospin funnel and centrifuged at 800 rpm for 5 min. Then the slides were stained with May-Grünwald-Giemsa (Sigma Aldrich) and the percentages of monocytes and macrophages were determined under a light microscope.
Determination of miR-155 and miR-125b expression levelsThe levels of miR-155 and miR-125b expression were measured by TaqMan® Small RNA Assays (Applied Biosystems, Foster City, CA, USA), using RNU48 as the reference gene. Briefly, total RNA was reversely transcribed using a specific looped primer, and reverse transcription quantitative PCR (qRT-PCR) was conducted using the standard TaqMan miRNA assay protocol. Amplification of miRNA by qRT-PCR was implemented by a LightCycler® 480 PCR System (Roche Molecular System, Pleasanton, CA, USA). The experiments were performed in triplicate. The relative expression of miRs normalized on the reference gene was calculated using the 2-ΔΔCT method.
Determination of macrophage function-related gene expression profileTo investigate the gene expression profile after PMA treatment, qRT-PCR was performed. Briefly, total RNA was extracted from each sample using a Hybrid-R™ RNA Isolation Kit (GeneAll, Seoul, Korea) according to the manufacturer’s instructions. The expression levels of TNFα, BACH1, CEBPβ and IRF4 genes were determined using two-step qRT-PCR. Reverse transcription was performed using a High Capacity RNA-to-cDNA kit (Applied Biosystems). SYBR Premix EX TaqTM (Takara Bio, Shiga, Japan) with the specific primers, TNFα: forward primer, 5’-CGAGTGACAAGCCTGTAGC-3’ reverse primer, 5’-GGTGTGGGTGAGGAGCACAT-3’, BACH-1: forward primer, 5’- TTCATGCTTCTGTTCAGCCAA-3’, reverse primer, 5’- GGCACTGAGAAGCAGG ATCTTT-3’, CEBPβ: forward primer, 5’- GACAAGCACAGCGACGAGTA-3’, reverse primer, 5’- AGCTGCTCCACCTTCTTCTG-3’, IRF4: forward primer, 5’- TCCGCCAGTGGCTGATCGAC-3’, reverse primer, GAPDH: forward primer, 5’- GAAGGTGAAGGTCGGAGTC-3’, 5’- reverse primer, GAAGATGGTGATGGGATTTC -3’ was used during the real-time PCR step followed by 40 cycles of denaturation at 95°C for 3 sec, annealing at 60°C for 30 sec, and finally the melt curve stage at 95°C for 30 sec, 60°C for 1 min, and 95°C for 30 sec. The amplification of target genes with GAPDH as an internal control was performed using a LightCycler® 480 PCR System (Roche Molecular System).
Phagocytic activity of PMA-treated U937 cells to yeastsThe 3x105 cells of U937 were treated with 0, 10, 25, 50, 100, and 200 nM of PMA in 500 µL of RPMI-1640 supplemented with 10.0% FBS and incubated at 37°C, 5.0% CO2 for 48 hr in a 24-well plate. The 5 x 107 of yeast cells (Saccharomyces cerevisiae) were stained with 20 µM of carboxy fluorescein diacetate succinimidyl ester (CFSE) (Molecular Probes-Invitrogen) in phosphate buffer saline (PBS) for 30 min before co-culture with PMA-treated U937 in the macrophage: yeast ratio at 1: 8 for 2 hr at 37°C with 5% CO2. PMA-untreated U937 cells were used as a control. Non-adherent cells were then washed off with PBS for 3 times. The adhered cells were scratched with a pipette tip and then stained with biotin-streptavidin PE antibody (eBioscience) conjugated anti-CD14. Phagocytic activity as double positive fluorescence (CD14+/CSFE+) was detected by flow cytometry (BD FACSCalibur™, BD Bioscience). The percentage and mean fluorescent intensity (MFI) of CD14+/CSFE+ cells were analyzed by Cell Quest software (Becton Dickinson). The experiments were performed in triplicate.
Statistical analysisThe data were analyzed using means ± standard deviations (SD). One-way ANOVA or Student’s t test was used to calculate statistical significance. [Statistical package for the Social Sciences (SPSSVR) version 13; SPSS Inc., Chicago, IL, USA]. A p-value less than 0.05 was considered statistically significant.
Cell proliferation and viability were determined by trypan blue staining after incubation with the five concentrations of PMA, 10, 25, 50, 100, and 200 nM, for 24 and 48 hr. The study found that the cell numbers which represented cell proliferation were not significantly different among the five concentrations in either the 24- or 48-hr treatments. The numbers of U937 monocyte-derived cells after 48 hr of PMA treatment were 3.56 ± 0.31 x 105, 3.42 ± 0.48 x 105, 3.46 ± 0.62 x 105, 3.15 ± 0.88 x 105, and 3.00 ± 1.35 x 105 cells/mL for 10, 25, 50, 100, and 200 nM of PMA, respectively, and there were no significant differences among these five groups as shown in Fig. 1A. The percentages of trypan blue negative cells which were considered as viable cells of PMA-treated U937 were significant lower compared to untreated condition after 24 and 48 hr of treatments. Cell viability of U937 after treatment with 200 nM was significantly lower than that in the condition with 10, 25, 50 nM at 24 hr and 10 nM at 48 hr (Fig. 1B). The viability percentages of U937 monocyte-derived cells after 48 hr of PMA treatment were 82.73 ± 1.48, 79.53 ± 0.47, 75.78 ± 4.59, 72.16 ± 8.54, and 68.66 ± 9.31, respectively, as shown in Fig. 1B. Using bright field microscopy, we observed increasing numbers of adherent macrophages after PMA treatment for all five concentrations (Fig. 2A). The macrophage-like phenotype was characterized after May-Grünwald-Giemsa staining by changes in morphology including cell size, nuclear morphology, an increase in the cytoplasmic ratio and the formation of vacuolization. This study found morphological changes in the U937 cell line in both the nucleus and cytoplasm in every condition of PMA treatment (Fig. 2B). The percentages of U937 monocyte-derived macrophages after treatment with 10, 25, 50, 100, and 200 nM of PMA for 48 hr were 79.5 ± 1.5, 77.5 ± 2.5, 78.5 ± 1.5, 78.5 ± 2.5, and 79.5 ± 1.5, respectively, all of which were higher than in the 24-hr treatments (69.0 ± 1.0, 67.5 ± 0.5, 69.0 ± 4.0, 73.0 ± 1.0, and 71.0 ± 2, respectively). There were no statistically significant differences between the five concentrations of PMA after 48 hr of treatment (Fig. 2C). The percentages of CD14 positive cells after treatment with the five concentrations of PMA were 95.61 ± 2.73, 97.3 ± 1.30, 98.32 ± 0.58, 98.4 ± 0.59 and 96.54 ± 1.95, respectively, while, it was 36.39 ± 1.21% in PMA-untreated condition. These results indicated that the five concentrations of PMA had similar effects in induction of macrophage differentiation.
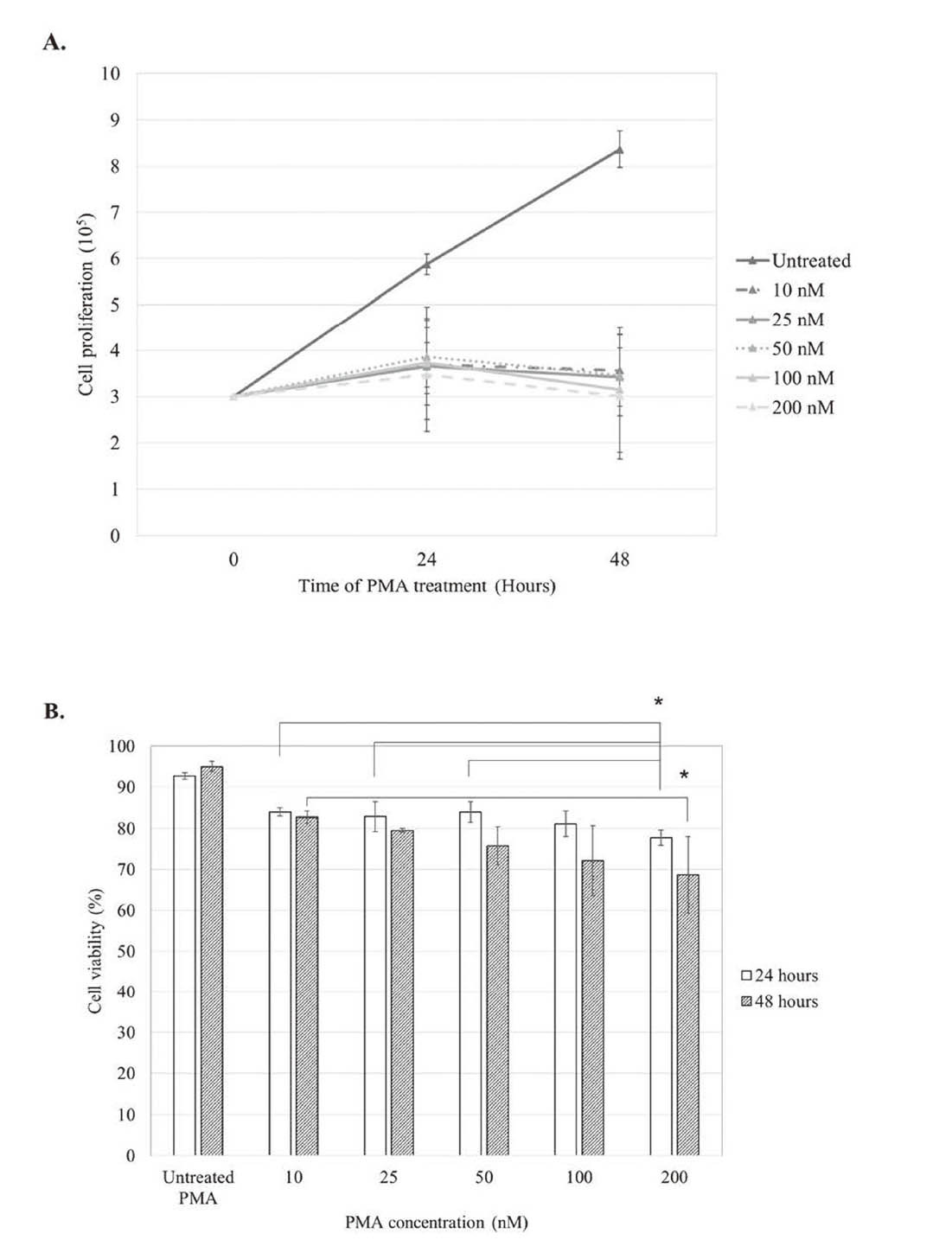
The effects of PMA concentrations on the proliferation and viability of U937-derived cells. The 3 x 105 cells of U937 were treated with 0, 10, 25, 50, 100, and 200 nM of PMA in 24 well-plates at 37°C, 5.0% CO2 for 24 and 48 hr. The cells were counted on hemocytometer using trypan blue dye with light microscope. Viable cells that could exclude the trypan blue appeared transparent while dead cells were stained blue. (A) Cell proliferation was assessed as the total number of cells. Cell proliferation after treatments with various concentrations of PMA was significantly lower than that in PMA-untreated cells at 24 and 48 hr. There was no significant difference of U937 cell proliferation among the five concentrations of PMA treatment at 24 and 48 hr (B) Cell viability was accounted as (number of live cells/total number of cells) × 100. Cell viability of PMA-untreated U937 was significant higher compared to other conditions after 24 and 48 hr of treatments. Cell viability of U937 after treatment with 200 nM was significantly lower than that in the condition with 10, 25, 50 nM at 24 hr and 10 nM at 48 hr. Data are shown as mean ± S.D. (n = 3/group). *: P-value < 0.05, significant differences in cell proliferation and viability were identified using one-way ANOVA.

The effects of PMA concentrations on the differentiation of U937-derived cells. The 3 x 105 cells of U937 were treated with 0, 10, 25, 50, 100, and 200 nM of PMA for 48 hr in 24-well plates. The cell morphology was studied using (A) brightfield microscopy (400X) and (B) May-Grünwald-Giemsa staining (1000X). (C) The percentages of monocytes and macrophages were determined under a light microscope after stained with May-Grünwald-Giemsa. At least 100 cells were counted on each slide. (D) The CD14 expression levels were measured with flow cytometry. The 1 x 105 cells from each condition were stained with biotin-streptavidin PE conjugated anti-CD14 before examined with flow cytometry. At least 10,000 cell signals were obtained in each sample and the data were analyzed by Cell Quest software.
The relative expression levels of miR-155 after treatments with 10, 25, 50, 100, and 200 nM of PMA were upregulated to 96.34 ± 0.67, 46.68 ± 3.07, 3.36 ± 0.06, 2.22 ± 0.12 and 2.19 ± 0.50, respectively, with p-values 3.3E-07, 2.8E-04, 1.8E-04, 2.6E-03, and 0.054, respectively, when compared to untreated cells (Fig. 3A). Our study found that the expression levels of miR-155 were increased in all five conditions of PMA in dose-reversal manner (Fig. 3A). The miR-155-related genes involved in macrophage functions including TNFα, BACH1, CEBPβ, were determined and compared to untreated cells. The relative expression levels of TNFα after being treated with 10, 25, 50, 100, and 200 nM of PMA were 4.74 ± 0.07, 2.81 ± 0.19, 2.30 ± 0.19, 2.88 ± 0.55 and 2.04 ± 0.30, and these upregulation levels were statistically significantly different with p-values of 2.9E-05, 0.002, 0.004, 0.02, and 0.02, respectively, when compared to untreated cells (Fig. 3B). The relative expression levels of BACH1 after 10, 25, 50, 100, and 200 nM of PMA treatment were 0.34 ± 0.05, 0.32 ± 0.03, 0.29 ± 0.001, 0.43 ± 0.03 and 0.32 ± 0.01, and these downregulation levels were statistically significantly different with p-values of 0.005, 0.01, 0.02, 0.03 and 0.004, respectively (Fig. 3C). The relative expression levels of CEBPβ were 0.72 ± 0.12, 0.79 ± 0.03, 0.65 ± 0.02, 0.72 ± 0.08 and 0.65 ± 0.04, respectively. (Fig. 3D). These downregulation levels were statistically significantly different with p-values of 0.047, 0.049, 0.002, 0.02, and 0.003, respectively. All together, these results indicated that a low concentration of PMA had a sufficient effect to upregulate the expression of miR-155 and that correlated with the increased expression of TNFα, and the decreased expression of BACH1, and CEBPβ, mimicking activated macrophages.

The expression of miR-155 (A) and its related genes involved in macrophage functions including TNFα (B), BACH1 (C), and CEBPβ (D) after 48 hr of 0, 10, 25, 50, 100, and 200 nM of PMA treatment. The levels of miR-155 expression were measured by TaqMan® Small RNA Assays, using RNU48 as the reference gene. The expression levels of TNFα, BACH1 and CEBPβ genes were determined using two-step qRT-PCR as mentioned in material and method. The relative expression of miR-155 and its related genes normalized on the reference gene was calculated using the 2-ΔΔCT method. The experiments were performed in triplicate. Data are shown as mean ± S.D., *: P-value < 0.05, significantly different from PMA-untreated U937 cells by Student’s t-test.
The relative expression levels of miR-125b after treatment with 10, 25, 50, 100, and 200 nM of PMA were 947.72 ± 78.6, 51.81 ± 11.3, 4.25 ± 0.36, 1.63 ± 0.06 and 1.21 ± 0.04, respectively. This indicated that the expression of miR-125b was significantly upregulated in PMA dose-reversal manner with p-value 0.007, 0.046, 3.07E-04, 0.008 and 0.03, respectively (Fig. 4A). The expression of IRF4 which its related gene involved in inflammatory inhibition was decreased to 0.24 ± 0.06, 0.31 ± 0.06, 0.21 ± 0.001, 0.34 ± 0.01 and 0.21 ± 0.05 with p-values 8.3E-05, 0.001, 1.1E-04, 1.1E-05, 4.7E-05 respectively.

The expression of miR-125b (A) and its macrophage function-related gene IRF4 (B) after 48 hr of 0, 10, 25, 50, 100, and 200 nM of PMA treatment. The levels of miR-125b expression were measured by TaqMan® Small RNA Assays, using RNU48 as the reference gene. The expression levels of IRF-4 gene were determined using two-step qRT-PCR as mentioned in material and method. The relative expression of miR-155 and its related genes normalized on the reference gene was calculated using the 2-ΔΔCT method. The experiments were performed in triplicate. Data are shown as mean ± S.D., *: P-value < 0.05, significantly different from PMA untreated U937 cells by Student’s t-test.
The phagocytic activity was determined after cocultured PMA-treated U937 cells with CFSE-stained yeast cells by measuring the percentage and MFI of CD14+/ CFSE+ (Fig. 5). The percentages of CD14+/ CFSE+ cells after treatment with the five concentrations of PMA were 48.81 ± 6.37, 44.25 ± 3.63, 44.73 ± 2.13, 36.43 ± 0.83, and 35.67 ± 1.31, respectively, while MFI were 563.08 ± 5.45, 473.52 ± 47.91, 446.60 ± 24.25, 378.40 ± 17.96 and 390.76 ± 11.06, respectively. Our result showed the increasing of phagocytic activity of U937-derived macrophages from the five concentrations of PMA treatments in dose-reversal manner when compared to PMA-untreated cells. The percentage and MFI of CD14+/CFSE+ PMA-untreated cells were 19.28 ± 2.70 and 300.83 ± 53.49, respectively.

The effects of PMA concentrations on the phagocytic activity of U937-derived cells. The 3 x 105 cells of U937 were treated with 0, 10, 25, 50, 100, and 200 nM of PMA for 48 hr in 24-well plates before co-cultured with saccharomyces cerevisiae in the macrophage:yeast ratio at 1:8. The phagocytic cells were detected using brightfield microscopy (400X) (A). The 1 x 105 cells from each condition were stained with biotin-streptavidin PE conjugated anti-CD14 before examined with flow cytometry. The percentage (B) and mean fluorescent intensity (MFI) (C) of CD14+/CSFE+ cells were measured with flow cytometry. At least 10,000 cell signals were obtained in each sample and the data were analyzed by Cell Quest software. The experiments were performed in triplicate. Data are shown as mean ± S.D., *: P-value < 0.05, significantly differences were identified using one-way ANOVA.
Our study found that the five usual concentrations of PMA had no significantly different effects in proliferation, viability and differentiation of U937-derived macrophages. However, the lower concentrations of PMA increased the higher expression levels of both miR-155 and miR-125b and phagocytic activity of macrophage.
A previous study reported increased expression of miR-155 and miR-125b in macrophages compared to monocytes of healthy human donors (Cobos Jiménez et al., 2014). A study showed that these two miRs were transiently under the direct control of NF-κB, which was a transcription factor that controlled transcription of DNA, cytokine production and cell survival (Tili et al., 2007). Interestingly, NF-kB activation has been reported as a mechanism of PMA that induces the differentiation of monocyte to macrophage (Krappmann et al., 2001). Therefore, NF-kB may be the key factor that regulates the expression levels of the two miRs after PMA treatment. Both miR-155 and miR-125b have been reported to be involved in various functions of macrophages by controlling the post-transcriptional regulation of a wide verity of proteins. A study showed that miR-155 probably directly targets transcript coding for Fas-associated death domain protein (FADD), IκB kinase ε (IKKε), and the receptor (TNFR superfamily)-interacting serine-threonine kinase 1 (Ripk1) while enhancing TNF-α translation in Raw 264.7 macrophage-like cells. The overexpression of miR-155 results in enhanced translation of TNF-α, probably by enhancing the stability of its transcript leading to increased production of TNF-α (Tili et al., 2007). The studies in miR-155 knockout (KO) mice demonstrated that miR-155 drives the inflammatory phenotype of M1-macrophages by regulating the expression of approximately 650 genes including IL-1β, IL-6, IL-12, and TNF-α (Jablonski et al., 2016). Our study examined the upregulation of miR-155 and TNF-α after PMA treatment. This result indicated that PMA had an effect in the induction of macrophage function involved in inflammatory response. Moreover, we found the suppression of the miR-155-targeted genes, BACH1 and CEBPβ, after PMA treatment. Previous studies found that the suppression of these two genes was involved in increasing phagocytic activity and inflammatory cytokine production of macrophages, respectively (He et al., 2009; Srinoun et al., 2017). The miR-125b is another miRNA that has been shown to be involve in macrophage functions. A study has revealed the increased level of miR-125b in macrophage compared to lymphoid, thymus and spleen cells. A previous study found that miR-125b was partly responsible for generating activated macrophages by reducing the levels of inflammatory response inhibitor, IRF4 (Chaudhuri et al., 2011; Honma et al., 2005; Negishi et al., 2005). However, to date, few studies have examined the regulators of IRF4 in macrophages. The miR-125b have also shown to response to the IFN-γ which is one of the M1 macrophage stimuli resulting in the increased the expression of MHC II, CD40, CD86, and CD80 activation markers (Chaudhuri et al., 2011). Our previous study found that the higher miR-125b expression in CD14-positive monocytic cells of β-thalassemia patients was positively correlated to phagocytic activity while negatively correlated to anemia severity (Kuno et al., 2019). In this study, both miR-155 and miR-125b were higher upregulated after treatment with the lower concentration of PMA and that correlated with the increasing of phagocytic activity of macrophage. The increased phagocytic activity of macrophage may be involved in many mechanisms including the increased production of TNF-α or macrophage colony-stimulating factor (M-CSF), and the increased recognition of phosphatidylserine (PS) or IgG on target cells by macrophage (Wiener et al., 1996, 1999; Banyatsuppasin et al., 2011; Wanachiwanawin et al., 1993; Chinprasertsuk et al., 1997). Therefore, from our results, at least the increased expression of TNF-α may be a mechanism that can activate phagocytic activity of U937-derived macrophage after PMA treatment.
Our result showed that the expression levels of miR-155 and miR-125b were upregulated after treatment with PMA in dose-reversal manner. The upregulation of miR-155 was correlated with increased expression of TNFα and decreased expression of BACH1 and CEBPβ, while the reduction of IRF4 was corelated to increased expression of miR-125b. However, the changing expression levels of the four macrophage functions related genes were not related to the expression levels of the two miRs. It may be because of the changes in expressions of the target genes may be affected by several factors. For example, the expression of IRF4 could be regulated by many factors such as GM-CSF or IL4 that can upregulate the expression of IRF4 by enhancing JMJD3 demethylase activity (Achuthan et al., 2016) or mTORC2 which can enhance the expression of IRF4 for M2 macrophage activation (Huang et al., 2016). These results indicated that the expression levels of a gene did not depend on the expression levels of a single factor or miR.
The results obtained in this study are important for the use of U937 cell lines as models for human macrophages, and keep the advantages of a cell line model which is easier to handle in performing experiments.
This work was supported by a “budget revenue” grant (MED600572S) of Prince of Songkla University and the Faculty of Medical Technology, and a “research credit” grant of the Faculty of Medicine, Prince of Songkla University. SK was supported by a scholarship from the Department of Biomedical Sciences, Faculty of Medicine, Prince of Songkla University. We would like to thanks professor Duncan R. Smith from Institute of Molecular Biosciences, Mahidol University for U937 cell line and Dr. Mingkwan Yingkajorn from microbiology unit, Faculty of Medicine, Prince of Songkla University for yeast isolation for phagocytic activity experiment.
Conflict of interestThe authors declare that there is no conflict of interest.