2025 Volume 50 Issue 6 Pages 263-272
2025 Volume 50 Issue 6 Pages 263-272
Cadmium (Cd) exposure through the respiratory system is associated with various respiratory disorders, including lung cancer. Circular RNAs (circRNAs) are increasingly recognized as critical regulators in carcinogenesis. This study employed high-throughput RNA sequencing and quantitative real-time PCR (qRT-PCR) to identify differentially expressed circRNAs in 16HBE cells (Cd-T) after 30 weeks of cadmium chloride (CdCl2, 5 μmol/L) exposure. Circ_0008272 knockdown inhibited migration, invasion, proliferation, and colony formation in Cd-T cells, whereas its overexpression enhanced these malignant phenotypes. Bioinformatics analyses and RNA-Protein Interaction Prediction (RPISeq) suggested that circ_0008272 promotes tumor-like behavior in Cd-T cells by modulating histone modification pathways. In conclusion, circ_0008272 acts as a tumor-promoting factor in the Cd-induced malignant transformation of 16HBE cells.
Cadmium (Cd) is a naturally occurring toxic metal that humans can absorb through multiple exposure pathways (Pan et al., 2024; Tucovic et al., 2018; Zheng et al., 2024). Cadmium exposure is strongly associated with multiple respiratory diseases, including asthma, increased pneumonia mortality, reduced lung function, emphysema, chronic obstructive pulmonary disease, and lung cancer (Brindhadevi et al., 2023; Chen et al., 2023; Park et al., 2020; Torén et al., 2019; Yang et al., 2019b; Zheng et al., 2021). The progression of cancer due to chemical exposure involves complex cellular interactions and disruptions in key biological pathways (Lu et al., 2024). Elucidating these molecular mechanisms is essential for evaluating cancer risk.
Circular RNAs (circRNAs), a unique class of non-coding RNAs, are resistant to degradation by RNA exonucleases due to their covalently closed circular structure, which endows them with distinct biological properties (Yang et al., 2019a; Zhou et al., 2021). Recent studies highlight the regulatory role of circRNAs in various malignancies, such as gastric, colon, ovarian, and lung cancers. For instance, circ_0006089 promotes gastric cancer progression by regulating NRP1 expression via miR-217 (Zhou et al., 2024), while circ_0017552 enhances proliferation and inhibits apoptosis in colon cancer through NET1 upregulation (Liu et al., 2024). In ovarian cancer, circFAM188A drives cell proliferation and invasion by interacting with miR-670-3p (Yong et al., 2024), and in lung cancer, circFBXW7 suppresses cancer stem cell renewal via the circFBXW7-185AA pathway (Li et al., 2023). Despite their established role in cancer progression, the expression patterns of circRNAs during chemical carcinogenesis remain poorly understood. A comprehensive investigation into the functions of differentially expressed circRNAs is crucial for elucidating the mechanisms of chemical-induced lung carcinogenesis. These findings could facilitate the identification of biomarkers for early detection of chemical-induced malignancy.
This study establishes a model of Cd-induced malignant transformation in human bronchial epithelial cells (16HBE) and identifies circ_0008272 as a novel circRNA involved in this process. By examining its functional role and predicting its regulatory network, the study provides foundational data for future molecular research into cadmium-induced carcinogenesis.
This study utilized Cd-induced malignant transformed cells (Cd-T) along with established lung cancer cell lines (A549, H1299, and H460) and immortalized human bronchial epithelial cells (16HBE). Cell lines were obtained from the Guangzhou Institute of Respiratory Diseases (Guangzhou, China). Lung cancer and Cd-T cells were cultured in RPMI-1640 medium (Gibco, New York, USA) supplemented with 10% fetal bovine serum (FBS) (Zhejiang Tianhang Biotechnology Co., Ltd., China) and 1% penicillin/streptomycin (Gibco, New York, USA). In contrast, 16HBE cells were cultured in MEM (Gibco, New York, USA) enriched with 10% FBS and 1% penicillin/streptomycin at 37°C in a 5% CO2 environment. Upon reaching 50%-60% confluence, 16HBE cells were treated with 5 μM CdCl2 (China Sigma Co., Ltd., Beijing, China). After 24 hr, the medium was discarded and cells subcultured. This Cd exposure was repeated for 30 weeks until malignant transformation was observed.
RNA extraction and qRT-PCRTotal RNA was extracted from the cells using TRIzol reagent (Invitrogen, Carlsbad, USA). cDNA was synthesized with the GoScriptTM Reverse Transcription System (Promega, Madison, USA). Quantitative real-time PCR (qRT-PCR) was conducted using the GoTaq® qPCR Master Mix kit for qRT-PCR. All qRT-PCR reactions were replicated using the Applied Biosystems (AB, Foster City, USA), and gene expression was quantified by the 2-ΔΔCt method, with GAPDH serving as the internal control. The sequences of the primers utilized are detailed in Supplementary Table S1.
RNase R treatmentA total of 1000 ng of RNA was used for both the control and experimental groups. In the experimental group, RNA was treated with 3 U/μg of RNase R (Epicentre, Madison, USA) at 37°C for 10 min. The stability of circ_0008272 and CTCF mRNA was then evaluated using qRT-PCR.
Separation of cytoplasmic and nuclear fractionsNuclear and cytoplasmic RNA fractions were separated using the Ambion™ PARIS™ Extraction Kit (Invitrogen, USA) following the manufacturer’s instructions. Fractionated 16HBE cells were analyzed for circ_0008272 expression using qRT-PCR. CTCF and U6 served as control markers for nuclear and cytoplasmic localization, respectively.
Fluorescence in situ hybridization (FISH)The circ_0008272-specific probe was synthesized using the Fish Kit (Sangon Biotech, China) according to the manufacturer’s protocol. After pre-hybridization, the hybridization buffer containing the anti-circ_0008272 probe was applied overnight at 37°C. Nuclear staining was performed using DAPI. Cell climbing slices were visualized and imaged using a Leica laser confocal microscope. The probe sequence for circ_0008272 is detailed in Supplementary Table S2.
Cell transfectionShort hairpin RNA (shRNA) and overexpression plasmids specific to circ_0008272 were synthesized by Gene Pharma (Shanghai, China). Cells were seeded into 6-well plates, and transfection was conducted at 70% confluency using Lipofectamine 3000 transfection reagent (Invitrogen, USA) following the manufacturer’s instructions. The sequences used are provided in Supplementary Table S3.
EdU assayCell proliferation was assessed using the EdU staining proliferation kit (RiboBio, China). Log-phase cells were seeded into 96-well plates. Each well was treated with 100 µL of EdU-supplemented medium and incubated for 2 hr. After labeling, cells were fixed with 4% paraformaldehyde, rinsed with PBS, and permeabilized using 0.5% Triton X-100. Following permeabilization, cells were stained with 1× Apollo for 30 min in the dark and treated with reagent F for an additional 30 min. Stained cells were visualized under fluorescence microscopy, and cell counts were subsequently determined.
Soft agar colony formation assayCell proliferation in non-adherent conditions was assessed using the soft agar colony formation assay. First, 1.2% and 0.6% low-melting agarose were prepared. The 1.2% agarose was mixed with 2×MEM medium (1:1) and added to 6-well plates as a bottom layer. Subsequently, 3×103 cells were mixed with 0.6% agarose and 2×MEM medium (1:1), overlaid on the bottom layer, and allowed to solidify. The plates were incubated at 37°C in a 5% CO2 incubator for 2 weeks. Colony formation was observed and recorded under a microscope.
Colony formation assayCells were seeded in 6-well plates at a density of 500 cells per well and cultured at 37°C in a 5% CO2 incubator for two weeks. Colonies were fixed with 4% paraformaldehyde and stained using 1% crystal violet. Colonies were observed, counted under a microscope, and the colony formation rate was compared between groups.
Cell scratch assayConfluent cells in 6-well plates were scratched using a sterile pipette tip. Three parallel guide lines were drawn on the underside of each 6-well plate. Cells were inoculated into the 6-well plates and cultured overnight to achieve confluency. Scratches were made using a sterile 200 µL pipette tip, creating a wound. Wells were rinsed with PBS to remove detached cells, followed by the addition of 2 mL of serum-free medium. Plates were incubated for 24 hr, and wound width was measured at 0 and 24 hr to calculate the healing rate using ImageJ:
Mobility = (0 hr width - 24 hr width) / (0 hr width) × 100%
Transwell assayCell invasion was assessed by diluting Matrigel in serum-free medium at a 6:1 ratio, with 50 µL of the diluted gel added to the upper chamber of Transwell inserts. A total of 4×104 cells were seeded into the upper chamber with serum-free medium, while 600 µL of medium containing 20% FBS was added to the lower chamber as a chemoattractant. Cells were incubated at 37°C in a 5% CO2 incubator for 24 hr. After incubation, cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Residual cells in the upper chamber were removed using a cotton swab. Images of eight random fields were captured using a microscope, and cells were counted with ImageJ software.
RNA-seq and data processingThe RNA-seq assay was conducted in accordance with the protocol provided by Personalbio (Shanghai, China). 16HBE cells were exposed to 5 μmol/L CdCl2 for 30 weeks, after which differentially expressed genes were identified through RNA-seq. Total RNA was extracted using the Trizol (Invitrogen, USA) reagent. Subsequent to this, Personalbio took charge of the RNA purification, library preparation, and sequencing procedures. Raw sequencing reads underwent filtering to yield clean reads, which were then aligned to the reference genome using HISAT. DESeq2 analysis was employed to identify the differentially expressed genes (P < 0.05, |log2FC| > 1.0, FDR < 0.05).
Plasmids and cloningThe circ_0008272 overexpression plasmid and the control empty vector pcDNA3.1-GFP were both custom-designed and synthesized by Gene Pharma (Suzhou, China). The circ_0008272 mature sequence overexpression plasmid was constructed by synthesizing a 319-bp fragment containing the exon sequence flanked by a about 60~90-bp intronic regions with complementary Alu repeats (Gene Pharma, China). pcDNA3.1(+) served as the cloning vector, and the overexpression sequence of circ_0008272 was circularized through the cloning sites BamHI and EcoRI, followed by enzyme digestion for 2 hr at 37°C. Subsequently, the digested products were purified and recovered by agarose gel electrophoresis, and the linearized target fragment was ligated to the vector using T4 DNA ligase (Fermentas, USA) for 2 hr at 22°C. Finally, the ligation mixture was transformed into competent cells TOP10 using the calcium chloride method, and clones were screened on plates containing ampicillin. The plasmid extraction was performed according to the instructions of the Plasmid Mini Kit (Tiangen Biotech, Beijing, China). 200 µL of the bacterial culture corresponding to the positive clone was sent for sequencing by Gene Pharma (Suzhou, China), and the remaining bacterial culture was preserved in glycerol. After confirming that the sequencing results matched the target gene sequence without errors, the preserved glycerol stock was used to inoculate LB broth for large-scale plasmid extraction, thereby obtaining a sufficient amount of recombinant plasmid.
Statistical analysisStatistical analyses were performed using SPSS 26.0, and graphical representation was generated with GraphPad Prism 8.0. All experiments were performed in triplicate, and data were presented as mean ± SD. An unpaired t-test was used for two-group comparisons, while one-way ANOVA was applied for comparisons involving more than two groups. A p-value <0.05 was considered statistically significant and denoted as *P < 0.05, **P < 0.01, and ***P < 0.001.
In this study, 16HBE cells were exposed to 5 μmol/L CdCl2 for 30 weeks, followed by RNA sequencing. RNA sequencing identified 20 significantly differentially expressed circRNAs (P < 0.05, |log2FC| > 1.0, FDR < 0.05). Among these, three downregulated circRNAs (circ_0007788, circ_0004243, and circ_0004210) and three upregulated circRNAs (circ_0005611, circ_0008272, and circ_0008599) were selected for further validation. qRT-PCR results confirmed that circ_0007788, circ_0004243, and circ_0004210 were downregulated, while circ_0005611, circ_0008272, and circ_0008599 were upregulated in Cd-T cells (Fig. 1A). Among these circRNAs, circ_0008272 exhibited the most significant differential expression. Moreover, the expression level of circ_0008272 in Cd-T, A549, H1299, and H460 cells was higher than 16HBE cells at the control level (Fig. 1B). Additionally, the mRNA expression level of CTCF, the parental gene of circ_0008272, was significantly elevated in Cd-T cells compared to that in 16HBE cells (Fig. S1).

Circ_0008272 is highly expressed in Cd-T cells and lung cancer cell lines. (A) qRT-PCR verification of 3 up-regulated circRNAs and 3 down-regulated circRNAs. (B) Circ_0008272 mRNA expression levels in Cd-T, A549, H1299 and H460 cells. *P < 0.05, **P < 0.01, *** P < 0.001.
Circ_0008272 comprises two exons (9–10) located at chr16:67628369–667629533 (Fig. 2A). Since circRNAs are resistant to RNase R digestion, RNase R treatment was performed to validate the expression of circ_0008272 in 16HBE cells. Results showed that circ_0008272 expression remained stable, whereas the linear RNA transcript (CTCF) was significantly degraded (Fig. 2B). Specific forward and reverse primers were designed for qRT-PCR analysis using complementary DNA (cDNA) and genomic DNA (gDNA) from 16HBE cells. Agarose gel electrophoresis confirmed that circ_0008272 could only be amplified in cDNA, not in gDNA (Fig. 2C). To determine its subcellular localization, cytoplasmic and nuclear fractions were analyzed, revealing that circ_0008272 primarily resides in the cytoplasm, with GAPDH and U6 serving as cytoplasmic and nuclear references, respectively (Fig. 2D). FISH assays confirmed these results, demonstrating that circ_0008272 predominantly localizes in the cytoplasm, where its 5’-FAM-labeled probes emitted green fluorescence, and nuclei stained with DAPI appeared blue (Fig. 2E). These findings indicate that circ_0008272 is a stable circRNA localized primarily in the cytoplasm.
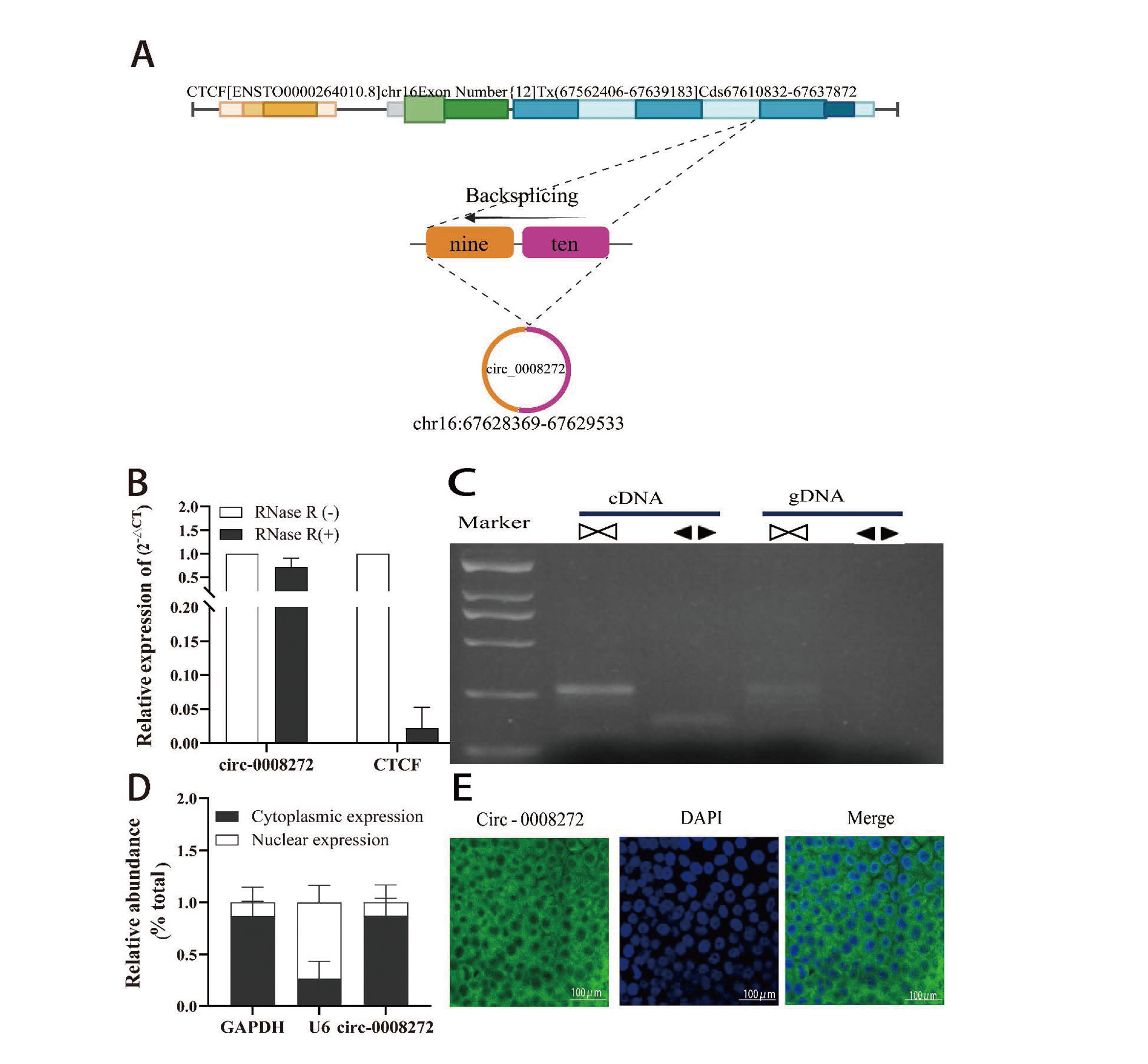
Circ_0008272 exhibits a stable circular structure and is located in the cytoplasm. (A) Schematic illustration showed the genomic loci of CTCF gene and hsa_circ_0008272. (B) qRT-PCR results showed circ_0008272 and linear CTCF mRNA expression levels in Cd-T cells treated with or without RNase R. (C) Agarose gel electrophoresis verification of circ_0008272 presence. Convergent primers amplified circ_0008272 from cDNA but not from gDNA. (D) Subcellular localization analysis of circ_0008272 in the cytoplasm and nucleus of Cd-T cells was analyzed by qRT-PCR. (E) FISH detection showed the cellular localization of circ_0008272 in Cd-T cells. *P < 0.05, **P < 0.01, *** P < 0.001.
To elucidate the role of circ_0008272 in Cd-T cell malignant transformation, two shRNAs (circ_0008272-S1 and circ_0008272-S2) and a negative control (NC) were transfected into Cd-T cells. The shRNAs reduced circ_0008272 expression by 63.1% and 40.2%, respectively (Fig. 3A). Circ_0008272-S1 was selected for subsequent experiments due to its higher interference efficiency. Additionally, an overexpression vector for circ_0008272 (OE) was transfected into Cd-T cells, achieving a notable level of expression (Fig. 3B). Anchorage-independent growth assays revealed that circ_0008272 knockdown suppressed the cells anchorage independent growth, a hallmark of malignant transformation, whereas circ_0008272 overexpression promoted this capacity (Fig.3C,D). Migration assays showed that circ_0008272-S1 markedly inhibited cell migration compared to the NC group, while circ_0008272-OE significantly enhanced migration (Fig. 3E, F). Similarly, Transwell invasion assays demonstrated that circ_0008272-S1 reduced invasion, whereas circ_0008272-OE increased invasive capacity (Fig. 3G, H). EdU assays and colony formation assays further confirmed that circ_0008272-S1 reduced cell proliferation, whereas circ_0008272-OE enhanced it (Fig. 3I-K). Collectively, these findings suggest that circ_0008272 promotes the malignant transformation of Cd-T cells.

Circ_0008272 promotion of malignant transformation in Cd-T cells. (A) qRT-PCR detection of circ_0008272-shRNA knockdown efficiency. (B) qRT-PCR validation of circ_0008272 overexpression efficiency. (C, D) Soft agar colony formation assay evaluated anchorage-independent growth of Cd-T cells with circ_0008272 knockdown and overexpression. (E, F) Cell scratch assay was used to examine the effects of circ_0008272 knockdown and overexpression on the migration of Cd-T cells. (G, H) Transwell invasion assay was used to detect the effects of circ_0008272 knockdown and overexpression on the invasive abilities of Cd-T cells. (I, J) EdU assay evaluated the impact of circ_0008272 knockdown and overexpression on Cd-T cells proliferation. (K, L) Colony formation assay assessed proliferation capacity of Cd-T cells with circ_0008272 knockdown and overexpression. *P < 0.05, **P < 0.01, *** P < 0.001.
To explore the functional mechanisms of circ_0008272, Gene Ontology (GO) analysis was performed on mRNA pathways associated with its parental gene, CTCF. The analysis identified 213 mRNA pathways, with 10 pathways significantly enriched (p < 0.05), including chromatin insulator sequence binding, DNA methylation/demethylation, histone modification, and related processes (Fig. 4A, B). To further explore the protein-protein interaction (PPI) network of CTCF, we employed the STRING database to identify interaction partners with a minimum interaction score threshold of > 0.15. The PPI network was then visualized utilizing the Cytoscape software (http://www.cytoscape.org/), where each node symbolized a distinct gene, and the edges depicted their interactions. Through this network, we found that EP300, SUZ12, NPM1 and YY1 emerged as the central elements of this intricate gene interaction network and were related to histone modification (Fig. 4C). Binding affinities between circ_0008272 and SMYD family proteins were predicted using the RNA-Protein Interaction Prediction (RPISeq) platform. The Random Forest (RF) and Support Vector Machine (SVM) classifiers predicted significant interactions between circ_0008272 and SMYD1 (RF: 0.7, SVM: 0.91), SMYD2 (RF: 0.6, SVM: 0.62), SMYD3 (RF: 0.7, SVM: 0.63), and SMYD5 (RF: 0.8, SVM: 0.44) (Fig. 4C). Visualization using Cytoscape revealed network interactions, suggesting that circ_0008272 may contribute to Cd-induced malignant transformation of 16HBE cells through histone modification.

Bioinformatics analysis of circ_0008272-associated network. (A, B) Top 10 enriched GO pathways and GO terms across different biological processes. (C) Protein-protein interaction network of CTCF and other proteins. (D) RPISeq analysis of the binding ability of circ_0008272 with the SMYD family.
The early stages of cancer development often involve the malignant transformation of normal cells, resulting in abnormal proliferation, migration, and other changes in cellular behavior. Prolonged exposure to carcinogens enhances cellular proliferation and differentiation, causing metabolic dysregulation and eventual malignant transformation (Gruenert et al., 1995). Since most human tumors originate from epithelial cells, in vitro carcinogen-induced transformation models of epithelial cells have been extensively used to investigate the mechanisms of carcinogenesis. For instance, bronchial epithelial cells (16HBE, BEAS-2B) exposed to carcinogens, such as cigarette smoke extract, arsenite, carbon black nanoparticles, and Benzo[a]pyrene (BaP), in long-term, low-dose experiments, have been used to establish malignant transformation models (Chen et al., 2024; Wu et al., 2024; Zhang et al., 2024a; Zhang et al., 2024b). A study revealed that cigarettes contain 0.5−1 μg of Cd, half of which can enter the body and accumulate in the lungs (Pinto et al., 2017). Adult lung capacity ranges from 2000 to 5000 mL (McDonough et al., 2015), with an average of 20 cigarettes smoked per day (Dai et al., 2022). The annual cumulative Cd intake reaches 2737.5 μg (0.75 μg × 20 cigarettes / day × 365 days), resulting in a cellular exposure concentration of 547.5 μg/L–1368.75 μg/L (2737.5 μg/5L, 2737.5 μg/2L), equivalent to 4.88 μM-12.21 μM. Similarly, our research demonstrated that prolonged exposure of bronchial epithelial cells (16HBE) to 5 μM Cd-chloride for 30 weeks resulted in malignant transformation (Wang et al., 2024). Using this Cd-induced transformation model, this study further investigated the role of cadmium in promoting malignant transformation.
CircRNAs, as key epigenetic regulators, have gained significant attention due to their structural stability and diverse functions. It has been proven that circRNAs exhibit resistance to RNase R degradation (Yu et al., 2018). This study confirmed the circular structure of circ_0008272, as it remained resistant to RNase R digestion, unlike the linear RNA transcript (CTCF), which was degraded. Aberrant circRNA expression has been implicated in malignant transformation. For example, silencing circ_0025373 in 16HBE cells significantly enhances their transformation, anchorage-independent growth, proliferation and migration (Zhang et al., 2024a), while circ_0087385 upregulation accelerates malignant transformation in response to BaP exposure (Zhang et al., 2024b). Conversely, circCIMT overexpression suppresses proliferation and invasion in Cd-transformed BEAS-2B cells (Chen et al., 2023). Our findings suggest that circ_0008272 acts as a tumor-promoting factor in Cd-induced transformation.
CircRNAs regulate gene expression through diverse mechanisms, including interactions with parental genes, transcription factors, RNA-binding proteins, and miRNAs (Cong et al., 2024; Hashemi et al., 2024; Shen et al., 2024). Notably, the parental gene CTCF of circ_0008272 was expressed at a higher level in Cd-T cells than in 16HBE cells, suggesting a strong correlation between the elevated expression of CTCF mRNA and the abundant presence of circ_0008272. Nevertheless, the precise mechanisms underlying this association warrant further investigation. YY1 can affect the methylation status of histones by interacting with histone modification enzymes, thereby regulating gene expression (Lewerissa et al., 2024). EP300, as a histone acetyltransferase, is specifically recruited to certain locations to acetylate the H3K27 locus in collaboration with TFAP2β, thereby facilitating the proliferation of neuroblastoma cells (Weinert et al., 2018). Interestingly, we discovered that genes involved in histone modifications, such as EP300, SUZ12, NPM1 and YY1, are linked to the parental gene CTCF of circ_0008272. SMYD family members, pivotal in cancer proliferation and metastasis, modify histones through methylation, acetylation, and other processes (Han et al., 2024). For instance, SMYD2 acetylates H3K36me2, promoting pancreatic cancer cell proliferation (Xu et al., 2024), while SMYD3 enhances gastric cancer growth by downregulating EMP1 via histone methylation (Zeng et al., 2023). Targeting histone-modifying activities of SMYD proteins may represent a promising therapeutic strategy. This study revealed that circ_0008272 interacts with SMYD1, SMYD2, SMYD3, and SMYD5, potentially driving malignant transformation through histone modification.
In conclusion, circ_0008272 promotes proliferation, migration, invasion, and anchorage-independent growth in cadmium-exposed cells. The interaction network identified in this study provides insights into potential regulatory mechanisms. However, these findings are based primarily on bioinformatic predictions. Future research should include extensive in vitro and in vivo experiments to validate the proposed mechanisms and clarify the specific regulatory roles of circ_0008272.
This work was financially supported by the National Natural Science Foundation of China (82160630 and 82373624 to HL) and the Natural Science Foundation of Inner Mongolia (2023LHMS08010 to HL).
Supplementary dataSupplementary data will be made available in the online version of the paper.
Conflict of interestThe authors declare that there is no conflict of interest.