2023 年 12 巻 1 号 p. A0129
2023 年 12 巻 1 号 p. A0129
Cancer metabolic variability has a significant impact on both diagnosis and treatment outcomes. The discovery of novel biological indicators and metabolic dysregulation, can significantly rely on comprehension of the modified metabolism in cancer, is a research focus. Tissue histology is a critical feature in the diagnostic testing of many ailments, such as cancer. To assess the surgical margin of the tumour on patients, frozen section histology is a tedious, laborious, and typically arbitrary method. Concurrent monitoring of ion images in tissues facilitated by the latest advancements in mass spectrometry imaging (MSI) is far more efficient than optical tissue image analysis utilized in conventional histopathology examination. This article focuses on the “desorption electrospray ionization (DESI)-MSI” technique’s most recent advancements and uses in cancer research. DESI-MSI can provide wealthy information based on the variances in metabolites and lipids in normal and cancerous tissues by acquiring ion images of the lipid and metabolite variances on biopsy samples. As opposed to a systematic review, this article offers a synopsis of the most widely employed cutting-edge DESI-MSI techniques in cancer research.
Approximately 10 million people worldwide die from cancer each year, which renders it as one of the leading causes of fatalities and illnesses globally. Cancer is still a difficult and fatal disease, in part because of its heterogeneous nature, despite the constant development of new therapies and treatments. For instance, the high inter- and intratumoral heterogeneity of malignancies like lung cancer, breast cancer, and glioma necessitates more individualized treatment.1) Cancer’s heterogeneity is greatly influenced by the complexity of the disease’s etiology and pathophysiology. Cancer is frequently brought on by accumulating genetic mutations and changes to the epigenome.1–3) However, environmental factors that contribute to carcinogenesis include specific diets, obesity, and persistent inflammation.4–6) As a result, the precise root cause of an illness could involve a range of associations among environmental and genetic components and could vary greatly between patients. The metabolic distinctions between cancer and normal cells within the tumor microenvironment are frequently blamed for poor treatment outcomes.7) To better understand the biology of cancer and develop more effective treatments, research on the varied characteristics of cancer tissues is essential. One of the most glaring characteristics of tumor tissues is the change in metabolic processes. Therefore, the study of regional metabolomics has been useful in determining the genesis, traits, and vulnerabilities of many malignancies. In this review, the promise of mass spectrometry imaging (MSI) as a contemporary molecular visualization technique with intriguing potential in cancer studies is addressed.
With the advent of new modalities and sophistication during the past 40 years, there remains a constant development in healthcare imaging techniques, which has had a significant influence on image-guided assessment and treatments.8) None of these methods (magnetic resonance imaging, positron emission tomography, or computed tomography) on tissue samples at an appropriate high resolution can directly determine the pathological traits associated with the disease subtype. Although significantly beneficial data can be acquired, tissue categorization with a microscope in histopathology can frequently become challenging and unreliable by frozen specimens, mimics, and discrepancies in tissue morphology.9) In addition, a faulty biopsy analysis often contributes to a botched surgical procedure by leaving perilous cells around the margin of the sample that was excised; this has been associated with a number of cancer variants, increasing regional recurrence and lowering overall longevity. Because of the requirement for speedy, precise diagnosis of cancer, the discipline of molecular medicine has grown increasingly dependent on biopsy specimen analysis.9)
By detecting the m/z of different molecular categories across tissue specimens, MSI, as an analytical approach, can offer spatially defined chemical data.9) The tissue surface is scanned in two dimensions during the imaging process, and then the mass spectrometric (MS) data are recorded pixel by pixel (Fig. 1). As a result, a significant amount of mass spectrum information is gathered throughout the tissue surface; therefore, any detected ion signals from chemical species can be paired with their spatial information to create their ion images.10) Due to the spatial mapping capabilities of ion signal, MSI can produce a large collection of ion images during a single scan. This makes it possible to simultaneously visualize (10–200 µm spatial resolution) the spread of hundreds of species from both inside and outside the tissue specimen, which can help with finding new biomarkers, the diagnosis of diseases, the development of new therapies, and pharmaceutical studies.11)
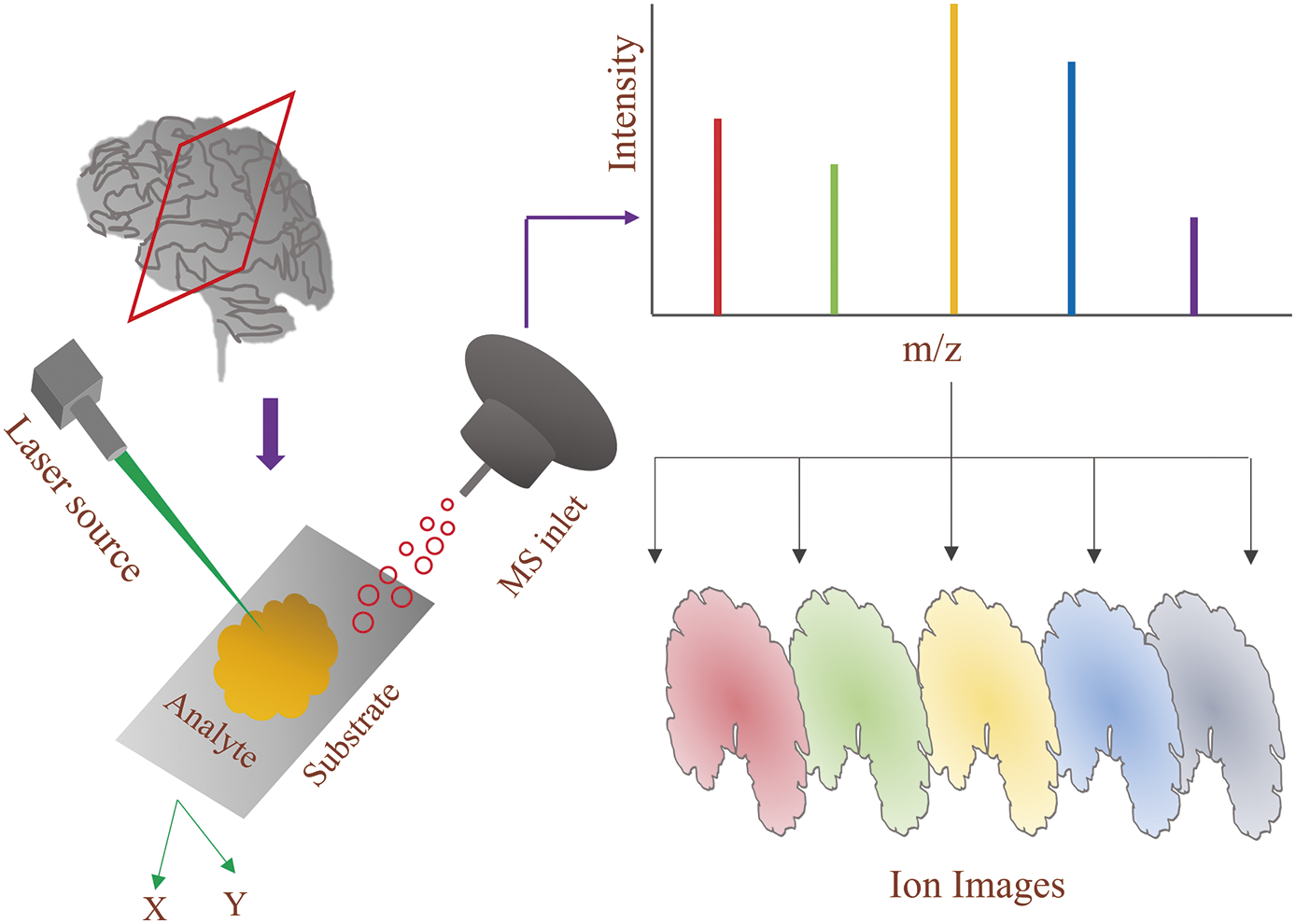
The effectiveness and applicability of MSI are normally assessed by considering two variables: (i) m/z spectrum and (ii) spatial resolution, for the investigation of several molecular groups in multiple tissues as well as diverse biological conditions. The desorption process determines the dimension of the probe, the layout of the mass detection system, and, ultimately, how the material to be examined is prepared.12) The MSI method’s spatial resolution, which is usually used alternately with pixel resolution in the discontinuous microprobe mode, is determined by the between points distance between every point where an MS experiment is acquired in line with the already established acquisition pattern.13) A wide range of biomolecules (proteins, peptides, etc.) in sophisticated biological tissues may be thoroughly image analyzed using MSI since it delivers the great molecular sensitivity that MS can offer.
The DESI-MSI technique (Fig. 2)14) visualizes small molecules in tissues that have been resected14) or unresected15) in a living organism. DESI-MSI is preferable to all comparable ionization procedures (secondary ion mass spectrometry, matrix-assisted laser desorption/ionization, etc.) since it may function in ambient circumstances and requires less sample preparation beforehand. DESI-MSI technique is also simple to operate since only an inexpensive solvent spray is required and does not need a vacuum environment for ionization. The DESI probe can instantly capture molecular fingerprints from live organs, tissues, etc.14) Typically, a tissue piece having an overall thickness of roughly 10–15 µm is used for the DESI-MSI. To electrospray a solvent at a high voltage while using N2 as the nebulizing gas, this tissue section was subjected to a stream of fast moving charged microdroplets.16,17) As a result of the solvent droplet saturating the tissue surface, the biochemical components contained in the tissue were dissolved. Like the ESI technique, the desolvation method utilizes continuous solvent removal and Coulomb fission of the subsidiary tiny droplets containing the analyte species to produce ionic substances in the gas phase.16) The bioimaging experiment hits the tissue surface with a spray of charged microdroplets while collecting mass measurements across the specimen and scanning it in both the horizontal and vertical dimensions. The biochemical makeup of the tissue can be mapped using the two-dimensional image made using the pixel-to-pixel MS spectra.18)

Chromatographic methods have been used in studies to identify widely known metabolites and variations in metabolic processes associated with several diseases, including hepatocellular carcinoma,19) neurodegenerative disease,20) and diabetes.21) However, since they need to have the molecules previously removed from the tissues, they are unable to preserve the spatial information of those metabolites.22)
Since spatial metabolomics preserves the spatial information of metabolites, it is essential to examine an array of small molecules simultaneously and thoroughly. A vast majority of studies focusing on spatial metabolomics employed MSI and ionization techniques to produce images of metabolite distribution.22) In contrast to conventional staining techniques, as a label-free methodology MSI can simultaneously detect many molecules.23) The alteration of metabolic pathways is a key feature of tumor tissues. Therefore, studying regional metabolomics has improved research into the genesis, traits, and vulnerabilities of various malignancies. The analysis of the diverse characteristics of cancer tissues is not possible due to the absence of spatial data in the metabolites.22) A new understanding of the metabolic reprogramming brought on by tumors has been provided using various MSI techniques in recent studies. The key findings of empirical research studies that have supported the value of DESI-MSI for visualizing metabolite and lipid distributions on human tissues in the context of cancer pathology will be summarized in this paper. In this review, the promise of MSI as a contemporary molecular visualization technique with intriguing potential in cancer studies is addressed. Key empirical research, cancer kinds, metabolites, metabolism processes, and clinical significance are listed in Table 1.
| Author | Diagnosis | Metabolites | Representative ions (m/z) | MP | CR | % Agt |
|---|---|---|---|---|---|---|
| Morse et al.117) | Prostate cancer | Fatty acids and phospholipids | LysoPE (16:0) (452.7), LysoPE (18:1) (478.2), PI (37:6) (867.5), PCh (O-40:2), PC (P-40:1) (826.7), FA 22:4 (331.2) | Krebs cycle | Prostate cancer metabolism and surgical guidance | 97.0 |
| Tamura et al118) | RCC | Lipids and fatty acids | Azelaic acid (187.1), (FA [16:1]) (253.2) (palmitoleic acid), (FA [18:2]) (279.2) (linoleic acid), (FA [18:1]) (281.2) (oleic acid) |
Warburg effect | Therapeutic strategy for targeting cancer cell metabolism | NR |
| Santoro et al.79) | Breast cancer | Lipids | Taurine (124.0), uric acid (167.0), GPLs (PE [P-16:0/22:6, 746.5], PS [38:3], m/z 812.5) |
De novo lipogenesis | Breast cancer pathogenesis | NR |
| Abbassi-Ghadi et al.108) | Esophageal adenocarcinoma |
Glycerol Phospholipids |
PG 36:4 (769.5), PG 38:6 (793.5), PG 40:8 (817.5), PI 34:1 (835.5) |
De novo lipogenesis | Early diagnosis of cancer | NR |
| Bensussan et al.119) | Adenocarcinoma and SCC | Glycerol Phospholipids |
FA (20:4) (303.2), PG (36:2) (773.5), PI (34:1) (835.5), PS (36:1) (885.5) |
NR | Classification of ADC and SCC subtypes |
87.5 |
| Zhang et al.45) | Renal oncocytoma vs. RCC | Free fatty acids (FFA) and monoradylglycerolipids (MG) | Ascorbic acid (175.0), hexose (m/z 215.0), FA (18:1) (281.2), FA (20:4) (303.2), CL (723.4 and 737.4) | NR | Discrimination of renal tumors | 99.5 |
| Vijayalakshmi et al.34) | RCC | Small metabolites, fatty acids, and lipids | N-acetyl glutamate (187.0), 2-hydroxy butyrate (103.0), creatinine (112.9) |
Metabolic alterations | Surgical margin assessment | 88.0 |
| Song et al.61) | Oral cancer | Amino acids, carbohydrates, glycerolipids, GPLs, and sphingolipids | Putrescine (89.1), betaine (156.0), phosphocholine (259.9), linoleic acid (317.1), hypoxanthine (175.0) | Arginine/proline metabolism, histidine metabolism | Satisfy the need for point-of-care testing | 88.9 |
| Fala et al.120) | Murine lymphoma | Metabolites | Pyruvate and lactate | Pyruvate metabolism | Investigate pyruvate delivery and lactate labeling | NR |
| Song et al.88) | TNBC | Metabolites | Arginine (175.1), lysine (147.1), N,N-dimethyl arginine (203.1), N,N,N-trimethyl lysine (189.1) | NR | Rapid diagnosis of TNBC | 88.8 |
| Strittmatter et al.115) | Pancreatic cancer | Gemcitabine | Ceralasertib (451.3), 2′,2′-difluorodeoxycytidine (302.0), 2′,2′-difluorodeoxyuridine (299.0) | Gemcitabine metabolism | Visualizing drug metabolites | NR |
| Kaufmann et al.121) | CRC | Fatty acids | Phosphatidylserines (810.5, 812.535, 766.536), PEs (726.5) | De novo lipogenesis | CRC diagnosis and prognosis | 97.0 |
| Vaysse et al.63) | Oral cavity cancer | Metabolites | Ether-PE(O-16:1/18:2) (698.5) | OSCC keratinization | In vivo tissue recognition | 96.8 |
| Huang et al.122) | Thyroid tumor | Amino acids, fatty acids, nucleotides, and lipids | Arachidonic acid (303.2), PIs (885.5), lysophospholipids (568.3) |
Pathogenesis mechanisms | Improve diagnosis accuracy | 93.0 |
| Mondal et al.99) | Breast cancer | Lipids, fatty acids | PC, PE, Cer, SM, DG | De novo lipogenesis | Therapeutic and diagnostic developments | 100.0 |
| Zhan et al.123) | Brain tumor | Small metabolites | Glutamine, hexcer (d42:2) | Endogenous metabolic pathways | Tumor research | NR |
| Aramaki et al.98) | Luminal breast cancer | Lipids and fatty acids | PC, TG, phosphatidylethanolamine, SM, and Cer | Cancer metabolism | Describe heterogeneity of cancer | NR |
% Agt, percent agreement; ADC, adenocarcinoma; Cer, ceramide; CL, cardiolipin; CR, clinical relevance; CRC, colorectal cancer; DG, diacylglycerol; FA, fatty acid; GPL, glycerophospholipid; Lyso PE, lyso-phosphatidylethanolamine; MP, metabolic pathway; NR, not reported; OSCC, oral squamous cell carcinoma; PC, phosphatidylcholine; PCh, phosphatidylcholines; PE, glycerophosphoethanolamine; PG, glycerophosphoglycerol; PI, glycerophosphoinositol; PS, glycerophosphoserine; SCC, squamous cell carcinoma; SM, sphingomyelin; TG, triglyceride; TNBC, triple-negative breast cancer.
More than ninety percent of renal patients with cancer who have been documented had renal cell carcinoma (RCC).24) RCC develops from proximal renal tubular epithelial cells. Over 140,000 kidney cancer deaths occurred in 2015, with an estimated 430,000 new cases of the disease being diagnosed worldwide.25) The prognosis of individuals with metastatic RCC has been shown to be greatly improved by effective comprehensive interventions, including immunological checkpoint inhibitors and tyrosine kinase antagonists.26–29) The choice of the most effective therapeutics for specific individuals, however, is not backed by sufficient evidence. As a result, new biomarkers are necessary for RCC early diagnosis in patients. Additionally, researching molecules that can indicate how a disease will develop or end helps find new biomarkers and enhances clinical outcomes.
According to metabolomics research, the immune system, nutrient depletion, and hypoxia all have an impact on cancer cell survival and proliferation.30) High levels of triacylglycerol and/or cholesterol ester have been discovered in the tumor tissue of clear cell renal cell carcinoma (ccRCC), the most common histological subgroup that accounts for approximately 70% of the different RCC types.31) Phosphatidylethanolamine upregulation caused by ethanolamine inhibits the proliferation of RCC cells in vivo.32) As opposed with traditional MS techniques, including liquid chromatographic procedures, which are widely employed for the screening of tiny biomolecules, MSI offers the benefit of permitting the direct investigation of associations between pathological observations.33) Additionally, DESI-MSI analysis enables the classification of tumor subclasses based on lipid profiles.
Vijayalakshmi et al. employed DESI-MSI to diagnose and predict clear ccRCC at a molecular level.34) Cross-sectional imaging has contributed to an increase in the prevalence of Stage 1 RCC, which has led to a rise in the use of nephron-sparing management strategies such as partial nephrectomy (PN) and ablation procedures.35) Positive surgical margins (PSMs) are more common in PN individuals with small renal masses and advanced RCC patients.36,37) Even though the impact of PSMs on longevity in RCC remains under discussion, empirical investigations have shown that they have a significant connection to greater frequencies of local relapse and a decrease in longevity irrespective of various other factors.38–41) Core biopsies’ frozen components may be utilized during surgery to confirm the diagnosis prior to performing ablative procedures.42) Intraoperative frozen sections’ (IFSs’) low utilization is a result of both their lengthy processing times and comparatively poor accuracy. In all, 23 frozen specimens of ccRCC underwent DESI-MSI. A cohort of 17 pairs of normal and tumor tissues was examined independently. The tissues underwent histopathologic analysis.34)
The authors noted variations in the abundance and distribution of small metabolites associated with the TCA cycle in the m/z 50–200 range, reflecting previously known altered metabolism in ccRCC.34) ccRCC showed an increased concentration of lactate, glucose, glutamate, N-acetyl glutamate, and 2-hydroxybutyrate compared to healthy tissue and a lower concentration of creatinine, fumarate, and succinate.34) The identified metabolites are roughly in accordance with the known metabolic alterations in ccRCC spurred on by alterations in hypoxia-sensing pathways brought on by functional inactivation of the von Hippel–Lindau.30,43) Within the m/z 200–1,000 range, variations in peak abundance were noticeable. The ccRCC tissue exhibits elevated concentrations of glycerophosphoglycerol, glycerophosphoinositol (PI), and glycerophosphoserine (PS) compared to the normal tissue. Aminophospholipids like glycerophosphoethanolamine (PE) are less prevalent in the ccRCC tissue. Reduced PS and PE levels can have a significant impact on lipid trafficking across membranes and the asymmetry of membrane structure, which influences apoptosis and phagocytosis, two processes involved in programmed cell death.32,44) The study demonstrates the potential of DESI-MSI and spatial metabolomics to precisely differentiate healthy and malignant tissues.34)
In a related investigation, Zhang et al. utilized DESI-MSI to distinguish between healthy kidney tissues, RCC variants, and renal oncocytoma (RO).45) RO, a common benign kidney neoplasm that makes up 5–9% of all renal tumors, is most frequently found in men over 50.46) The diagnosis of RO frequently happens by chance during routine medical exams because it is asymptomatic. Early assessment of renal tumors can be accomplished through the use of radiological imaging techniques such as ultrasound,47) magnetic resonance imaging,48) computed tomography,49) and positron emission tomography.50) The ability of these diagnostic imaging procedures to reliably differentiate among malignant and benign tumors is limited, despite the fact that they are efficient at detecting and finding worrisome kidney malignancies. More renal tumors are accidentally discovered because of increased imaging use, which increases the likelihood of surgery.51) The sizable histologic and immunophenotypic overlap between RO and malignant chromophobe renal cell carcinoma (chRCC) makes it difficult to diagnose the difference between the two using cytological and histological evaluation, which is one of the reasons why preoperative core needle biopsies of renal lesions are not more widely used.52,53) Consequently, it is essential to distinguish ROs from RCCs, especially the closer related kinds, such as chRCCs, in order to ensure proper management of renal neoplasms in patients.45)
Utilizing a pair of normal kidney tissues, 2 specimens of RO tissue, and 2 samples of RCC, appropriate DESI-MSI images were acquired. The mass spectra of normal kidney samples were obtained from renal cortex tissues only to provide a fairer comparison given that ROs originate because of the distinctive molecular patterns found in healthy renal cortex and medulla, from the region of the renal cortex.45) In the abundances of ions m/z (391.2, 656.5, 737.4, 810.526, 835.5, and 863.561) while examining different kinds of tissues, patterns differed, but among two specimens of the same type, patterns were consistent. Except for m/z 737.4, all six of the presented ions have relatively high abundances in normal renal tissues.45) When comparing RCC and healthier kidney tissues, the RO samples had lower abundances of the molecules at PS (20:4_18:0), PI (34:1), and PI (36:1), but higher abundances of m/z 737.4 (Fig. 3). RCCs showed similar abundance to normal samples for ceramide (Cer, d40:1), m/z (810.5, 835.5), but lower abundance for m/z (392.0, 864.0) when comparing RO and healthy tissues.45) Overall pixel and patient accuracy for the different tissue types was 99.47%. Overall, the study shows that metabolic information gathered in conjunction with statistical methods enables the distinguishing of renal tumors and, as a result, has the potential to be applied in the healthcare context for improving the treatment of individuals who have renal tumors.45)

With a mortality rate of more than 50%, oral squamous cell carcinoma (OSCC) constitutes by far perhaps the most lethal cancer of the oral cavity. OSCC is a multistep neoplasia that begins as a benign oral epithelial hyperplasia and evolved to dysplasia and then carcinoma.54) Local recurrence is yet another factor contributing to treatment failure, even in OSCC patients who have undergone surgical resection.55) Currently, a visual examination of the mouth combined with histology is still the preferred method for identifying oral cancer.56,57) Despite the relatively easy access to the oral cavity, some asymptomatic cases during or after surgery can be challenging to observe. Therefore, creating a useful tool is widely desired for molecular detection in biological fluid. Even though traditional mass spectrometry methods (liquid,58) gas,59) and capillary electrophoresis60)-coupled MS) have found use in the biomedical and clinical fields, the methods necessitate time-consuming pretreatment of complex biological samples prior to analysis. When the use of such a sample size exceeds hundreds or thousands, restrictions are unavoidable and prevent platforms for salivary point-of-care tests because analysis takes at least a few weeks or months.
OSCC was diagnosed from saliva by Song et al. using conductive polymer-based paper spray ionization (CP-PS-MS) and DESI-MSI.61) Saliva specimens have been obtained from healthy individuals, individuals who had premalignant lesions, and individuals with OSCC in order to discover and authenticate dysregulated metabolites and uncover altered pathways in metabolism. Even though salivary metabolomics can typically represent the metabolic situation within the principal OSCC site, the metabolite concentration was eventually watered down to some degree due to continuous saliva secretion.62) By using DESI-MSI, metabolite indicators were reaffirmed at the principal tissue level. The metabolic profile of the saliva can indicate the emergence of oral cancer. Saliva has the potential to be used for the diagnosis of OSCC because many newly discovered metabolites were found to be strongly correlated with the expression levels of those metabolites within the tissues of the primary oncological site, the oral cavity.61) By group analysis with CP-PS-MS, it was discovered that in the precancerous stage, small metabolic changes begin with aminoacyl t-RNA biosynthesis; arginine, histidine, and proline metabolism; and lysine degradation. Metabolites implicated in the pathways were observed to be increasingly up- or downregulated when the tumor attained the aggressive stage. OSCC and precancerous tumors can be differentiated instantaneously from typical physical issues with over 87% precision using CP-PS-MS and DESI-MSI, as well as machine learning (ML).61)
During oral cavity cancer surgery, rapid evaporative ionization mass spectrometry (REIMS) can detect even a small proportion of tumor cells, as recently investigated by Vaysse et al.63) Due to its hemostatic characteristics, that are necessary for cauterizing the vascularized tissues in the oral cavity, electrocautery is often utilized as a surgical technique for resecting the oral cavity. One of the byproducts of this technique is electrosurgical vapor. Recent empirical investigations on the identification of normal tissues and intraoperative cancer using REIMS evaluation of these vapors have yielded encouraging results.64,65) Databases of tissue-specific metabolic parameters were created using REIMS analysis of cauterized tissue sections.
Eleven OSCC patients have been included in the study. In 185 REIMS, the tissue was divided according to its metabolic properties, and using cross-validation and multivariate statistical methods, the tissue categorization was contrasted to histopathology.63) REIMS results were aided by the implementation of DESI-MSI, which was utilized to determine tissue heterogeneity and was performed on six sections of the oral cavity. REIMS sensitivity was evaluated using a novel cell-based assay composed of various cell lines. REIMS correctly identified tumors and soft tissues in 96.8% of cases. Among 90% of myoblasts, REIMS was able to identify tumor cells with more than 80% sensitivity and specificity.63) The phosphatidylethanolamine PE(O-16:1/18:2) and cholesterol sulfate metabolic shifts widespread to both mucosal developmental processes and OSCC. Variations were emphasized by DESI-MSI, besides the unique metabolic properties of nerve properties (Fig. 4). During oral cavity cancer surgeries, the assessment of tissue differences using REIMS and DESI-MSI responsiveness with cell compositions described sensitive metabolic characteristics during in vivo tissue identification.63)

Breast cancer is the most prevalent kind of cancer in women, which is a complex group of illnesses with wide variations in their clinical manifestations and biologic aggressiveness. Advances in genomic profiling and a plethora of clinical studies of breast cancer have produced evidence for the presence of clinically practical molecular subgroups.66) The introduction of this molecular subtype into clinics has been facilitated, in part, by evaluation of hormonal receptors (ER and PR) and HER2 status.67) The Warburg effect, or metabolic reprogramming in cancer cells, was first identified almost a century ago, but interest in it has only recently grown again due to the hypothesis that it constitutes one of the traits that set cancer cells apart.68,69) De novo lipogenesis is involved in several cellular processes, including oxidative stress-induced cell death, chemotherapeutic agent uptake regulation, and the production of signaling molecules.70) De novo lipogenesis, a characteristic of aggressive cancers, is induced by genomic alterations in breast cancer, such as the deletion of chromosome 8p.71–74)
Breast cancer diagnosis and molecular subtyping are frequently done in clinics using histopathologic interpretation of hematoxylin and eosin (H&E) staining of tissue sections, immunohistochemistry (IHC) assays that are unique for ER and PR determination, and fluorescence in situ hybridization for the evaluation of HER2 gene amplification.70,75) H&E and IHC, despite being relatively straightforward techniques, are time-consuming experiments that are vulnerable to bias because of reaction conditions as well as information interpreting bias. With the help of DESI, researchers can study cancer cell metabolism with little sample pretreatment.18,76–78) DESI is a recent and significant advancement in ambient ionization techniques.
The chemical make-up of the breast cancer subtypes was characterized by DESI-MSI by Santoro et al.79) In addition to metabolomic characteristics at 200 µm, areas of focus included invasive breast cancer (IBC), ductal carcinoma in situ (DCIS), and adjacent benign tissue (ABT). Ions observed in IBC areas included sphingolipids, deprotonated glycerophospholipids (GPLs), and polyunsaturated fatty acids. Highly saturated lipids; antioxidant molecules such as taurine, uric acid, ascorbic acid, and glutathione; as well as ABT and IBC can be used to differentiate them. Furthermore, triple-negative and luminal B subtypes demonstrate more intricate lipid profiles when contrasted to luminal A and HER2 subgroups. DCIS and IBC were differentiated from one another based on cell signaling and apoptosis-related ions. In situ metabolomic results presented here contribute to our understanding of the pathogenesis of the cancer subtypes of breast cancer, healthy tissue, and DCIS.79)
According to Ahmad, in triple-negative breast cancer (TNBC), 20% of all occurrences of breast cancer are of the most severe and deadly form, which carries a significant risk of recurrence and distant metastases.80) Researchers have recently had the opportunity to examine enormous amounts of data at the molecular level, thanks to the advancement of omics techniques. This might make it easier to fully comprehend the molecular alterations and underlying processes involved in the growth of cancer.81,82) A range of factors must be carefully considered for an IVD tool to be effective in clinics: 1) Easily identifiable indicators with outstanding sensitivity and selectivity; 2) an easily accessible, appropriate biological material; and 3) innovative tool with reasonable test expenses, trustworthy test performance, and rapid result feedback.83)
Ambient ionization mass spectrometry (AIMS) has garnered a lot of interest in clinical metabolomics due to its distinctive advantages in direct measurement of lipids and metabolites from the bioanalyte under ambient conditions.84–86) One of the noteworthy AIMS methods, conductive polymer spray ionization (CPSI)-MS, has been utilized to assess the metabolic characteristics of biological fluids.87) Song et al. quickly extracted the metabolic signature from a ≤1-µL liquid extraction of the TNBC tissue that had been biopsied employing ML and CPSI-MS. The study employed a 4-step work process: 1) metabolite differentiation in TNBC tissue; 2) monitoring TNBC-affiliated metabolites in serum and developing metabolite-based ML models for swift serum monitoring and tissue detection, respectively; 3) pathway enrichment assessment to find prospective enzymes and carriers affiliated with the dysfunctional metabolites; and 4) expression verification of enzymes or carriers liable for the dysfunctional metabolites. The targeted metabolites were verified in tissues using DESI-MSI.88)
In all, 76 different metabolite indicators are acknowledged at the main site of carcinoma and can be satisfactorily tracked in serum. Rapid serum monitoring with 15- and 22-metabolites produces, when identified by CPSI-MS, tissue prognosis results with a sensitivity of 89% and a specificity of 90%, respectively. The initial verification of the upstream enzyme and carrier expression levels has now been completed. The cost-effectiveness of CPSI-MS technology makes it possible to quickly screen for, detect, and characterize TNBC metabolism reprogramming in clinical settings.88)
Although cancer cells have been characterized using cellular and genomic analysis to find care plans,89,90) cancer tissues have more distinctive features than the cellular component, making it feasible to categorize cancers more accurately if cancer tissues can be more meticulously described. Due to the wide range of carbon double bond configurations observed in lipid molecules, these molecules display a great deal of diversity, and this diversity represents the complex pathophysiology of cancer tissues in a significant manner.82,91) Additionally, it has been proposed that the tumor grade and diagnosis are linked to differences in the metabolism of a wide range of lipids in breast cancer, such as sphingomyelin (SM), phosphatidylcholine (PC), triglycerides (TGs), and phosphatidylethanolamine.91,92)
The genetics and phenotype of breast cancer can be categorized.93) Using next-generation sequencing, the diversity of genotypes has been investigated.94) Numerous genetic investigations have been widely employed in the treatment of breast cancer, despite the fact that gene mutations are less common in this type of cancer compared among other malignancies.95) The pathological technique for assessing cancer morphologies will not be deemed irrelevant irrespective of how advanced genomic approaches become; in fact, it will become more important since phenotypes may illustrate the impact of genetic anomalies. MSI can identify and display compounds on histological slices while preserving spatial information of the molecules. It is possible to determine the m/z of lipids and fatty acids using MSI. Furthermore, MSI investigation has shown that cancer tissues contain higher levels of specific fatty acids and lipids than normal tissues do.96,97)
To examine variation in luminal breast cancer biopsy specimens, Aramaki et al. performed cluster evaluation of MSI data based on the properties of lipid molecules and the degree of their expression.98) The clusters were composed of phosphatidylethanolamine, SM, PC, TGs, and Cer. It was discovered that the percentage of TGs and PC mainly correlated with the percentage of stroma and cancer on hematoxylin & eosin (HE) images. This group of lipids also differed from cluster to cluster in their carbon composition.98) This was in line with the premise that an upsurge in the total number of long-chain fatty acid synthesizing enzymes is brought on by cancer metabolism. If PC aggregates exhibit elevated carbon levels, that might be a sign of severe malignancy. According to these findings, malignancies for whom genetic evaluation simply is insufficient can potentially be categorized using phenotype heterogeneity in conjunction with lipidomics.98)
In a different study, Mondal et al. imaged fresh-frozen excision samples from 73 cancer patients who experienced lumpectomies using DESI-MSI. This included cancer and paired adjacent normal tissues.99) The results demonstrate a marked metabolic upregulation of diacylglycerol (DG), a lipid second carrier that activates protein kinase C to promote cancer growth (Fig. 5). Four different sn-1,2-diacylglycerols have been identified by the study.99) Using supervised ML on the complete dataset, diacylglycerols outperformed other lipid categories in successfully forecasting breast cancer, giving complete reliability in the validation data set. This accuracy considerably declined once DG signals were excluded from the ML process. Since DG is a possible oncomarker, DESI-MSI should complement the perioperative surgical pathology in lumpectomy.99)

Around 500,000 people die from esophageal cancer each year, making it the 8th most common cancer worldwide.100) Esophageal adenocarcinoma (EA), a variant of cancer, is most prevalent in the Western world and is connected to gastrointestinal acid reflux. Despite advances in comprehensive therapy, the 5-year mortality rate following therapy with therapeutic objective is still 30–35% and 5-year survivability throughout every phase is 14%.101) Therefore, new tools are required to encourage early diagnosis and efficient treatment. Lipids account for 70% of the human metabolome and have garnered little research focus, even though the relationship involving cancer and metabolism has been the subject of substantial research.102–105) Most lipids are locked away as GPLs in bilayer membranes. They are a desirable subject for biomarker studies due to their chemical and physical stabilities. Functionally, membrane properties and cellular signaling are impacted by GPL diversity. Phosphatidylglycerols (PGLs) have powerful and opposing effects on the proliferation of squamous cells,106) whereas phosphatidylinositols (PIs) mediate phosphatidylinositol-3 kinase (PI3K) signaling, one of the most frequently deregulated pathways in EA.107) Determining the specific species composition is necessary, although detecting GPLs may provide diagnostic indicators that are suitable for therapeutic use.
Abbassi-Ghadi et al. utilized DESI-MSI to contrast the typical squamous and EA GPL characteristics in an empirical investigation.108) The researchers additionally examined into the following: The development of the EA GPL profile was compared between normal, inflammatory, metaplastic, dysplastic, and neoplastic cell types, and these additionally functioned as an additional validation cohort. The underpinning genetics and related fatty acid pool served as the mechanistic foundation for these GPL markers. The presence of polyunsaturated PGLs with longer acyl chains was found to be elevated in EA specimens, among several other variations, with systematic enrichment in pre-malignant tissues. Fatty acid acyls’ properties were comparable to those of GPL acyls, and the genes encoding for fatty acid and GPL production were expressed at significantly higher levels. De novo lipogenesis is connected to the phospholipidome by inhibiting the carbon swap ACLY in EA cells.108)
Pancreatic ductal adenocarcinoma (PDAC) has a five-year survival rate of just 9%, making it one of the most fatal cancers. In a PDAC mouse model, accumulation of fatty acids was found to be correlated with PDAC initiation and metastasis,109,110) suggesting that lipid metabolism may be downregulated in PDAC. The phenotypic diversity and high heterogeneity of cancer cells are intimately associated with the pathogenesis of the disease.111) Delivery of drugs and response to therapy are thought to be constrained by dense fibrotic desmoplasia in PDAC because it prevents drugs from reaching the tumor cells.112–114)
Gemcitabine is a prodrug whose intracellular production of active metabolites makes it crucial to visualize the distribution of both the parent drug and its metabolites.115) Gemcitabine (dFdC) is an effective procedure for pancreatic cancer, but it is possible that it will not work because the tumor stroma blocks drug delivery to the cancer cells. Gemcitabine’s intratumor distribution (Fig. 6) and effect on PDAC by multidimensional imaging were developed by Strittmatter et al. in 2022.115) Numerous analytical techniques were used in addition to DESI-MSI to assess the local distribution and metabolic activity of gemcitabine in cancers from a genetically engineered mouse model of pancreatic cancer (KPC), allowing analogies between consequences in the tumor tissue and its microenvironment.115) With the aid of DESI-MSI, the spread of gemcitabine, its phosphorylated metabolites, and the dysregulated metabolite dFdU could be visualized.115) Distribution was contrasted with the Ataxia telangiectasia and RAD3-related kinase (ATR) inhibitor small molecule AZD6738. In contrast to the parent compound, gemcitabine metabolites were distributed differently within the tumor. While AZD6738 was discovered in untenable tumor areas, areas with plausible tumor cells had higher concentrations of dFdCMP, dFdCDP, and dFdCTP, which correlated with the spread of endogenous adenosine triphosphate (ATP), adenosine monophosphate (AMP), and adenosine diphosphate (ADP).115) This implies that gemcitabine’s active metabolites have access to the tumor cell. The method demonstrated that in KPC PDAC tumor tissue, higher proliferation sites were primarily correlated with the creation of active, phosphorylated dFdC metabolites and care-induced DNA damage.115)

From a laborious traditional chromatographic technique, empirical studies reviewed in the article provide an entirely novel method in assessing pathogenic pathways that is exceedingly effective, informative, and functional in real time. AIMS has radically altered the discipline of analytical chemistry by enabling researchers to study samples swiftly and directly in their native condition. With AIMS, sample preparation may be eliminated from workflows, sample processing times can be significantly reduced, and evaluation independent of the conventional laboratory environment has become a genuine option. The effectiveness of MSI as a tool to investigate the in situ spatial localization of molecular components has been demonstrated. In contrast to other widely used biochemical imaging methodologies, label-free screening is possible with MSI without the need for any prior knowledge of the potential target species. Even though MSI investigations are by nature untargeted, the process of preparing samples can be tailored for several various classes of intended biomolecules of relevance. This could provide more complex molecular information to connect the specialist domains of pathophysiology and metabolism in cells. In sharp contrast to the DNA, the metabolome linked to cancer is dynamic and greatly altered by inheritance, dietary habits, and medications. The potential for different results obtained through DESI-MSI investigations116) on each tumor model encourages more research in this rapidly evolving discipline.
The spatial data of the metabolites in numerous types of cancer needs to be further studied, as the field is still in the beginning stages. Such metabolite information may not only aid in the discovery of new biomarkers but also aid in the development of a thorough understanding of tumor pathophysiology. A key element for the effective implementation of this methodology is the discovery of metabolic biological indicators with spatially specified biochemical profiles connected to various diseases. Some believe that the recently developed AIMS approach will likely have a large positive impact since it can improve cancer ablation or therapy’s effectiveness and so aid in the widespread adoption of a therapy alternative with considerably fewer negative effects. It is necessary to develop new statistical techniques for rapidly acquiring detailed molecular information from ion images to enhance the diagnostic capacity provided by AIMS. AIMS must be evaluated on an extensive population of individuals with various kinds of malignancies before being implemented in routine pathological practice.
Future research may assess further data and employ novel strategies. For instance, it would be advantageous to connect metabolite distribution across the stages of malignancy once additional spatial metabolomics data on various cancer specimens become accessible. By contrasting the physiological and metabolic facets of cancer development, we can develop a greater understanding of the molecular mechanisms underlying tumor progression. Finally, merging information from the fields of omics (proteomics and spatial metabolomics) might just be an attractive step given that multi-omics investigation is a highly relevant area of research. By directly examining metabolic events in tissue specimens, MSI research has helped to comprehend the biological genesis and pathological characteristics of different carcinomas. The ongoing advancement of MSI and its expanded uses promise to reveal more about the molecular processes underlying diseases and disorders.
In light of all the experimental investigations establishing novel avenues in diagnostic molecular pathology, the discipline of clinical MS anticipates a shift from a slow and laborious widespread chromatographic approach to a simple and spatially resolved in situ MSI methodology that is specifically effective, instructive, and functional in real time. Each cancer model’s DESI-MSI study could yield distinct outcomes, opening the door to more research. It is projected that different tumors would have distinct molecular characteristics. The metabolome is also dynamic and significantly affected by a person’s diet, lifestyle, use of prescription medications, hereditary factors, etc., unlike the DNA of a cancer. Innovative statistical techniques for rapidly collecting complete molecular information from ion images will be needed to improve the diagnostic efficacy of DESI-MSI. It is necessary to perform this procedure on an adequate number of specimens from individuals who have a range of tumors to validate it prior to implementing it into a standard pathological practice. In addition to broadening the application of MSI to various tumor types, researchers can also keep investigating more minute spatial variations in metabolite distributions. Future research may very well be able to analyze novel details in metabolite distribution patterns with meticulous specimen preparation and ultrahigh-resolution and sensitive MSI methods. The development of swifter scanning techniques and ambient rapid ionization for MS will advance this nascent but remarkably appealing domain of spatial metabolomics soon.
The described study did not generate or analyze any data.
The authors are pleased to acknowledge the professors for their support who provided helpful criticism that helped to raise the manuscript’s overall quality.
The research was not supported by outside organizations.
The authors declare no potential conflicts of interest.
The study did not involve human or animal subjects.
Mass Spectrom (Tokyo) 2023; 12(1): A0129