2024 Volume 13 Issue 1 Pages A0142
2024 Volume 13 Issue 1 Pages A0142
Among the most typical posttranslational modifications is glycosylation, which often involves the covalent binding of an oligosaccharide (glycan) to either an asparagine (N-linked) or a serine/threonine (O-linked) residue. Studies imply that the N-glycan portion of a glycoprotein could serve as a particular disease biomarker rather than the protein itself because N-linked glycans have been widely recognized to evolve with the advancement of tumors and other diseases. N-glycans found on protein asparagine sites have been especially significant. Since N-glycans play clearly defined functions in the folding of proteins, cellular transport, and transmission of signals, modifications to them have been linked to several illnesses. However, because these N-glycans’ production is not template driven, they have a substantial morphological range, rendering it difficult to distinguish the species that are most relevant to biology and medicine using standard techniques. Mass spectrometry (MS) techniques have emerged as effective analytical tools for investigating the role of glycosylation in health and illness. This is due to developments in MS equipment, data collection, and sample handling techniques. By recording the spatial dimension of a glycan’s distribution in situ, mass spectrometry imaging (MSI) builds atop existing methods while offering added knowledge concerning the structure and functionality of biomolecules. In this review article, we address the current development of glycan MSI, starting with the most used tissue imaging techniques and ionization sources before proceeding on to a discussion on applications and concluding with implications for clinical research.
Glycosylation of proteins is a posttranslational modification that is crucial for biological functions such as cell–cell interaction, and morphological and modulatory processes.1) More specifically, glycoproteins play a role in the mechanisms of protein folding, molecular transportation, and secretion.2–4) Several natural and adaptive immune response regulatory mechanisms, such as cell trafficking, cell growth and differentiation, T and B cell receptor activity, antigen distribution, and antibody functioning and clearance, depend on protein and lipid glycosylation.5–7) Membrane-bound glycoproteins function as adhesive binding agents for host–host interactions, such as adhesion of cells, receptor activation, and extra cellular matrix (ECM) compounds, as well as host–pathogen connections, which allow pathogens such as viruses and bacteria to adhere to and enter mammalian cells.8,9) The technique of covalently attaching a sugar molecule (glycan) to an amino acid asparagine (N-linked) or serine/threonine (O-linked) domain is known as glycosylation. These sugars have the option of being joined as monomers or connected to form lengthy, complicated chains. Individual sugar components utilized to generate glycans happen to be absorbed entirely via the cell or produced through multiple metabolic processes and include, but are not constrained to, glucose (Glu), mannose (Man), fucose (Fuc), N-acetylglucosamine (GlcNAc), and N-acetylgalactosamine (GalNAc).10) Collectively, these chains of sugars create the glycocalyx, a thick sugar architecture that is found on the outermost layer of a cell.11)
N-glycans are biosynthesized in the endoplasmic reticulum (ER) as a dolichol-linked sugar base, passed on to nascent proteins during translation and then managed by several enzymes in the ER and Golgi apparatus as a component of the secretory process.12) The three primary N-glycan structural classes—oligo or hybrid, high Man, and complex—are the end products of this procedure. As a result of pathogenesis and as a direct regulator of cancer progression, N-glycosylation has been determined to be considerably modified in cancer.13) To assess N-glycosylation, numerous techniques have been successfully established.14) Lectins are frequently utilized for identifying N-glycans by immunohistochemistry (IHC), immunofluorescent, or colorimetric experiments for the investigation of tissues and surface-attached molecules. Lectins have relatively varied preferences and are typically specialized for adhering glycan structural components as opposed to certain glycan species, which restrict their application for exact glycan structural assessments. In the past decade, liquid chromatography combined with mass spectrometric methods has become a potent method for analyzing N-glycans. Although efficient, these techniques necessitate time-consuming sample processing stages that prevent the evaluation of considerable specimen batches in a practical amount of time in the context of prospective diagnostic or therapeutic applications. Although matrix-assisted laser desorption ionization (MALDI)-time of flight (TOF) mass spectrometry (MS) speeds up data collection, extensive purification and modification processes are still necessary before evaluation.15) The drawbacks of these earlier methods are addressed, and mass spectrometry imaging (MSI) for N-glycans also adds its own beneficial aspects.16,17) Timescales that are suitable for high rates of processing as well as clinical applications allow for the completion of increasingly complicated examinations into exact N-glycan structural connections. In this review article, we address the current development of glycan MSI, starting with the most used tissue imaging techniques before proceeding on to various substrates and categories of samples with possible implications for both clinical and academic studies. Table 1 presents a summary of the empirical studies, the nature of the ionization source and MS analyzer, tumor type, sample type, associated N-glycans identified, and supplementary techniques.
| Study | Tumor | Sample | Ionization | Analyzer | N-glycans associated* | ST |
| Vos et al.257) | Adenocarcinoma | Endoscopic mucosal (FFPE) | MALDI-MSI | TOF/TOF | Hex6HexNAc5, Hex6HexNAc5NeuAc1, Hex7HexNAc6 |
H&E stain |
| Black et al.258) | Liver cirrhosis | Serum | MALDI-MSI | FT-ICR | Hex3dHex1HexNAc4+Na, Hex5dHex1HexNAc4+Na, Hex4dHex1HexNAc4+Na | Anitbody array |
| Ochoa-Rios et al.180) | iCCA & HCC | Serum and iCCA/HCC tissue | MALDI-MSI | QTOF | Single fucosylated N-glycan, double fucosylated N-glycan, double fucosylated with a galactose N-glycan, tetraantennary branched N-glycan, high Man N-glycan | H&E stain |
| Ščupáková et al.63) | Breast cancer | Cancerous tissue | MALDI-MSI | TOF and FT-ICR | High-Man glycans, fucosylated and complex glycans | IHC and GE |
| Rebelo et al.201) | Neuroinflammation | Brain tissue section | MALDI-MSI | QTOF | Sialylated, fucosylated N-glycans and oligomannose N-glycans | PET, IHC, UPLC, RT-qPCR |
| Drake et al.259) | ccRCC | ccRCC tissue | MALDI-MSI | FT-ICR | Hex7HexNAc6Fuc1, Hex5HexNAc5Fuc1-3, Hex6HexNAc6Fuc1-3 | H&E stain |
| Blaschke et al.164) | Prostate cancer | Urine and prostatic fluid | MALDI-MSI | QTOF | Hex5HexNAc4NeuAc1, Hex5HexNAc4Fuc1NeuAc1, Hex7HexNAc6Fuc2 | HPLC, H&E stain |
| Boyaval et al.200) | Colorectal cancer | Tissue sections | MALDI-MSI | TOF/TOF | H9N2, H8N2, H7N2, H6N2, and H5N2 | H&E stain, CE-ESI-MS/MS |
| McDowell et al.163) | Pancreatic cancer | Pancreatic cancer FFPE tissues | MALDI-MSI | QTOF and FT-ICR | Hex6HexNAc5, Hex1HexNAc6, Hex9dHex1HexNAc8, Hex5dHex1HexNAc4, Hex7HexNAc6 | IHC, IF |
| Ochoa-Rios et al.180) | Nonalcoholic steatohepatitis | NAFLD/NASH tissue | MALDI-MSI | QTOF | Fucosylated glycan | H&E stain |
| Conroy et al.260) | Prostate cancer | Prostate cancer FFPE tissue | MALDI-MSI | TOF | High-Man glycans, fucosylated, bisecting, and sialylated glycans | NA |
| Grzeski et al.192) | OC | EOC FFPE tissue | MALDI-MSI | TOF | α2,6- and α2,3-sialylated N-glycans | H&E stain |
| Briggs et al.186) | OC | OC FFPE tissue | MALDI-MSI | TOF/TOF | (Hex)6+(Man)3(GlcNAc)2, (Hex)2 (HexNAc)2 (Deoxyhexose)1+(Man)3(GlcNAc)2 | H&E stain, PGC-LC-ESI-MS/MS |
| Heijs et al.50) | Neurological disorders | Cerebellum FFPE tissue | MALDI-2-MSI | TIMS-TOF | [M–H]– N-glycans | H&E stain |
| Conroy et al.261) | Prostate cancer | Prostate cancer FFPE tissue | MALDI-MSI | TOF | High Man, tri- and tetra-antennary glycans | NA |
| Pace et al.144) | Prostate cancer | Prostate Cancer FFPE tissue | IR-MALDESI-MSI | Orbitrap | Sialylated N-glycans | NA |
| Lee et al.262) | Osteoarthritis | Osteochondral FFPE tissue | MALDI-MSI | TOF/TOF | (Hex)4(HexNAc)3, (Hex)4(HexNAc)4, and (Hex)5(HexNAc)4 | H&E stain |
| Heijs et al.50) | MLS | FFPE MLS tissue | MALDI-MSI | TOF | Man N-glycans and tri- and tetra-antennary N-glycans | H&E stain |
| West et al.131) | Prostate cancer | Prostate FFPE tissue | MALDI-MSI | FT-ICR | Fucosylated N-glycans | SDS-PAGE |
| Boyaval et al.199) | Colorectal cancer | FFPE CRC tissue | MALDI-MSI | TOF/TOF | Hex7HexNAc2, Hex4dHex3HexNAc5, Hex5dHex1HexNAc4NeuAc | H&E stain |
| Delacourt et al.263) | HCC | FFPE tumor tissue | MALDI-MSI | TOF and FT-ICR | Hex6dHex1HexNAc6, Hex7dHex2HexNAc6,Hex7dHex3HexNAc6, Hex6dHex2HexNAc5 | H&E stain |
| Zhang et al.264) | Laryngeal tumor | FFPE human laryngeal cancer tissue | MALDI-MSI | TOF | Sialylated N-glycans | H&E stain |
| Angel et al.119) | Aortic valve stenosis | FFPE tissue | MALDI-MSI | FT-ICR | SA (NeuAc) N-glycans w | Lectin stain |
| Lu et al.162) | Prostate cancer | Tissues, cells, and biofluids | MALDI-MSI | QTOF | Alkyne-modified α2,3-linked SA N-glycans | H&E stain |
| Samal et al.222) | Neuro disorder | FFPE mouse brain tissue | IR-MALDESI-MSI | Orbitrap | N-sialoglycans | NA |
*Only representative metabolites are shown.
ccRCC, clear-cell renal cell carcinoma; CE, capillary electrophoresis; CRC, colorectal cancer; EOC, epithelial ovarian cancer; ESI, electrospray ionization; FFPE, formalin-fixed, paraffin-embedded; FT-ICR, Fourier transform ion cyclotron resonance; H&E, hematoxylin and eosin; HCC, hepatocellular carcinoma; HPLC, high performance liquid chromatography; iCCA, intrahepatic cholangiocarcinoma; IHC, immunohistochemistry; IR-MALDESI, infrared matrix-assisted laser desorption electrospray ionization; LC, liquid chromatography; MALDI, matrix-assisted laser desorption ionization; Man, mannose; MLS, myxoid liposarcoma; MS, mass spectrometry; MSI, mass spectrometry imaging; NA, not applicable; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; NeuAc, N-acetylneuraminic acid; OC, ovarian cancer; PET, positron emission tomography; PGC, porous graphitized carbon; QTOF, quadrupole time of flight; RILI, radiation-induced lung injury; RT-qPCR, real-time polymerase chain reaction; SA, supplementary techniques; SARS-CoV-2, severe-acute-respiratory-syndrome-related coronavirus; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; ST, supplementary techniques; TIMS, trapped ion mobility spectrometry; TOF, time of flight. UPLC, ultra performance liquid chromatography.
The spatial distribution of biological materials can be examined in situ using MSI, which has proven to be an extremely efficient technique for doing so. In comparison with other, better-known biochemical imaging methodologies, MSI offers label-free screening without necessitating a working understanding of the potential target species. Sample preparation can be tailored for various categorizations of the targeted molecules of interest, even though MSI evaluations are untargeted.18) As an ex vivo molecular visualization technique, MSI helps to further comprehend the biochemical processes happening in the tissue by highlighting key biomolecules. A common characteristic of many neurological disorders is molecular downregulation.19–22) Another aspect to bear in mind is the fact that MSI differs considerably from traditional MS-based omics techniques in that it examines how chemicals spread on the tissue in space rather than employing extracts of tissues or fluids.23) MSI has created new potential in the field of clinical MS by anticipating the switch from the time-consuming liquid chromatography (LC)-MS technology to an efficient and spatially resolved in situ procedure. As a result, MSI has created new possibilities for the field of clinical MS. In fact, it has been demonstrated that MSI is quite helpful for assessing the distribution of biomarkers in tissue sections to determine the boundaries of malignancy.24–27) The specimen surface is scanned in 2D for imaging before MS data are recorded pixel-by-pixel. As a result, a significant quantity of MS data is acquired throughout the sample surface, and any detected chemical species’ ion signals can be combined with their spatial data to produce their ion images.28) When structural markers in traditional histopathology are ambiguous, the evaluation of tissue metabolites usually offers more tangible information on the underlying condition.29) The MS-targeted technique differs from conventional molecular visualization using chemical discoloration, immunostaining, fluorescence, and radiolabeling in that it does not require prior knowledge of the species of interest to be utilized.29) Investigators are able to further closely investigate the chemical makeup of tissues with the aid of this MSI property in order to appreciate the functioning of multiple biochemical reactions that are crucial to living systems and identify any anomalies that might be related to them.29)
Consequently, the proliferation of multiple endogenous species in the tissue samples is continuously examined, which can aid in the discovery of biomarkers, the detection of diseases, and the development of innovative medicines.30) In order to assess the efficiency and suitability of MSI for the investigation of various molecular species in tissues, two factors are often prioritized: (i) spatial resolution and (ii) m/z range. The desorption process, which comprises the size of the probe, the layout of the mass detector, and most significantly, the sample preparation, has an impact on both variables.31) The several ionization processes employed in MSI, according to McDonnell and Heeren,32) are individually defined by complimentary benefits and drawbacks that focus on spatial resolution, mass accuracy and resolution, chemical sensitivity, specificity, and m/z range.32) When measuring gaseous ionic species in an MS, sample ionization is necessary to move the analyte from the condensed to the gas phase.32) A wide range of lipids, peptides, and proteins in intricate biological tissues may be thoroughly image analyzed using MSI since it delivers the great molecular specificity that MS can offer.
The mass range of N-glycan ions is quite broad, spanning from the most basic sugar chains (700 m/z) to configurations that are several times larger (4,000 m/z) for branched, fucosylated, and sialylated molecules, which is close to the maximum limit of detection with many existing MALDI-MSI systems.33) Reliable identification of the ionic species constitutes a topic that needs more research because there are numerous glycan species with uncertain makeup that transcend 4,000 m/z.33) The appearance of sulfate moieties in some species, isotopic overlap across structures, interference from the chemical matrices, and the development of sodium adducts connected to sialic acids (SAs) all add to the complexity.33) Although N-glycans are frequently identified in positive ion settings, there are some unique applications that are more appropriate for negative mode evaluation.34) To achieve these objectives, two general modes of MALDI-MS—the Fourier transform ion cyclotron resonance (FT-ICR) and TOF systems—each with distinct benefits and drawbacks—are generally utilized to identify discharged N-glycan ions from tissues. Recently, N-glycan MSI has been performed using instrument combinations that include a second MALDI setup (referred to as MALDI-2) and ion mobility spectrometry (IMS) paired with MALDI-quadrupole (Q)-TOF. Among the empirical studies reviewed, two of the most common instruments utilized were TOF and FT-ICR. MALDI-2 and trapped ion mobility spectrometry (TIMS)-TOF were used in limited capacities.
According to the fundamental theory of TOF, ions are separated depending on how long it takes them to pass down a flight path of a certain length and reach the detector.35) The fact that there is not any theoretical restriction to the maximum m/z detection limits of TOF devices is a benefit. Since m/z in such instruments inevitably depends on time spent in flight, lengthening the drift tube enables the identification of analytes with higher masses.36) Additionally, N-glycans from tissue homogenate are frequently assessed using TOF-MS by spotting on multisample target discs due to their higher processing capabilities. Despite less resolving power, the relatively brief time between the laser-ejection of N-glycan ions to their identification improves the number of pixels, which can be evaluated in a particular period, leading to the realistic viability of significantly higher spatial resolution evaluations (~5 μm).37,38)
Utilizing varying radiofrequency pulses and a strong magnetic field to excite N-glycan ions, ion-cyclotron resonance produces differential cyclotron resonance frequencies of the ion packets inside the detector that are correlated to their m/z.39–42) Opposed detector plates inside the ICR cell pick up this mixture of resonant frequencies as just one time-dependent current, which is subsequently deconvoluted into its individual frequencies by Fourier transformation. According to Bowman et al., FT-ICR instrument’s maximum resolving power is proportional to magnetic field strength.43) For high-mass analytical substances (2,000 m/z), however, its resolving power decreases without appropriate source changes.44)
While MALDI stands out among the empirical research under consideration as the preferred choice of ionization for the study of N-glycans, a few of the investigations used infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) as the ionization source.
In MS, MALDI is a soft ionization methodology that rapidly and efficiently ionizes a wide range of compounds.45) Detecting a wide range of polar compounds, particularly big macromolecules like proteins and peptides, has made MALDI (Fig. 1) a popular MS approach. The biological study that uses small sample sizes (few microliters) has been greatly impacted by the application of MALDI. When ionized by more traditional techniques, these molecules fragment excessively because they are typically fragile.45) When it comes to generating low molecular fragmentation upon ionization, MALDI and electrospray ionization (ESI) are comparable in nature. MALDI and ESI vary primarily in that the former usually yields ions with a net single charge, making it easier to determine the molecular mass of most molecules. It may, nevertheless, also make it more difficult to examine the biggest macromolecular proteins.45)
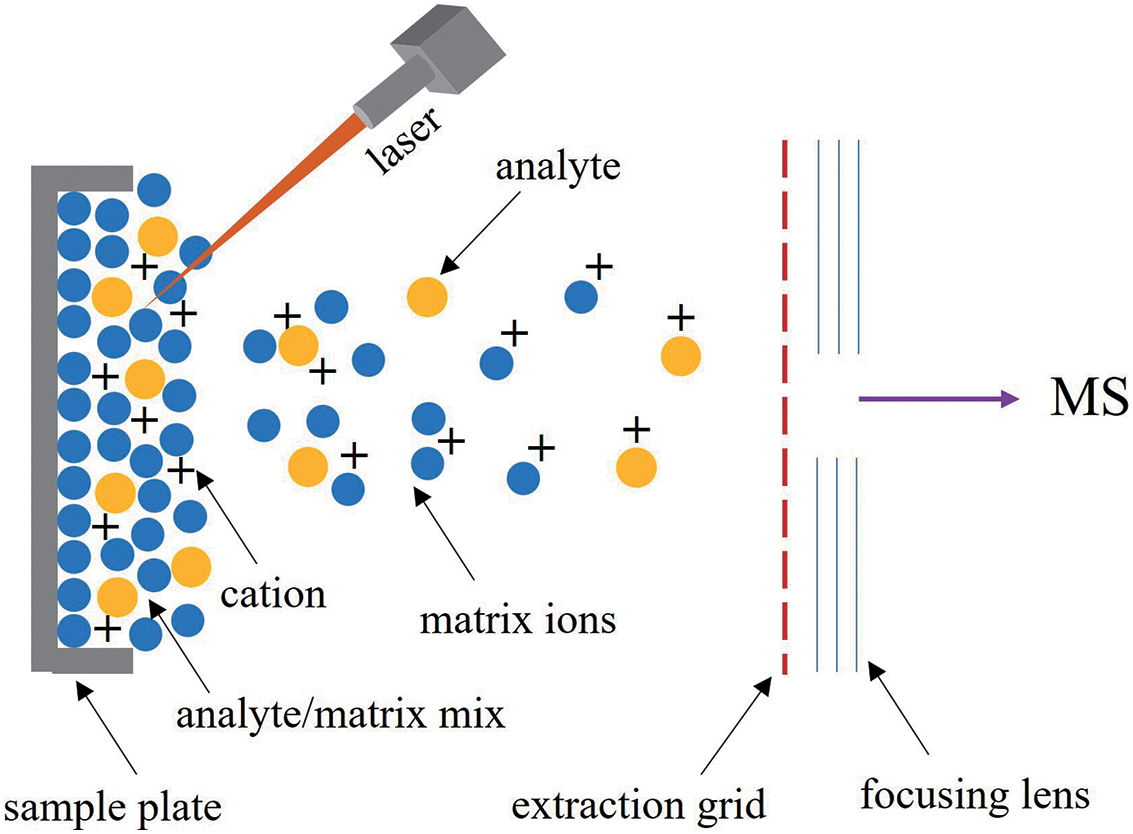
Three steps are involved in MALDI ionization. a) A suitable matrix material is combined with a sample before it is placed on to a metal plate. Crystals that are essential for effective ionization form on the sample material when the mixture dries. b) A pulsed laser beam impinges on the sample, producing desorption of the sample and matrix material. In the process, the matrix material decomposes into gas phase ionic species. c) In the hot cloud of ablated gases, the analyte molecules undergo protonation or deprotonation to get ionized.45) For mass analysis, the ions are subsequently accelerated into MS equipment. Due to several factors, including the matrix makeup, laser beam, and analyte dispersion in the sample, MALDI-MSI has a spatial resolution range of 5 to 50 μm.46)
The identification of analytes with extremely low abundances or high molar masses is a challenge shared by all MALDI techniques. This makes it more difficult to analyze mammalian N-glycans, which are created by including polylactosamine extensions.47) Ionization effectiveness and analyte breakdown in the MALDI source are significant factors causing this deficiency.47) Laser-induced ionization of analytes in a MALDI apparatus is comparatively ineffective when compared to alternative ionization schemes like ESI. This is particularly true for substrates with low proton affinity, such as carbohydrates, which ionize with an efficiency of 10–7–10–8 in MALDI experiments.48,49)
According to Heijs et al.,50) the analysis of N-glycans using a MALDI-2 instrument operating in the negative mode showed an order of magnitude more sensitive identification of N-glycans from brain tissue samples than did a typical MALDI platform operating in the positive mode.50) Tissue analysis at subcellular resolution is another option made possible by mass spectrometers equipped with MALDI-2. By using a transition-geometry MALDI-MS in conjunction with a secondary positronization laser to place the primary laser behind the tissue sample, single-cell analysis is made possible with pixel sizes of around 600 nm.51) Given that the volume of sample ejected into the instrument reduces proportionately with lower pixel size, secondary post-ionization is essential for producing an acceptable signal at these low rasters.52)
Glycan imaging has generally used a positive ionization approach since negative mode investigations have been less accurate in MALDI investigations.53,54) As a result, without the employment of extra chemical derivatization techniques throughout sample preparation, an ionization technique like ESI may be suitable for N-linked glycan imaging. IR-MALDESI achieves benefits from both soft ionization sources, including spatially correlated data from MALDI and ionization attributes from ESI, by combining both MALDI and ESI properties. High mass accuracy and resolving power produced by IR-MALDESI interfaced with Orbitrap MS are essential for the reliable investigation of biological molecules in untargeted MSI databases.53,54)
The IR-MALDESI (Fig. 2) method, which combines MALDI and ESI, operates in ambient settings, needs little sample preparation time, and provides the capacity to perform absolute metabolite measurement because a specific tissue volume is consistently ablated.55)

By combining charged electrospray droplets with the aerosol of ablated material, IR-MALDESI creates ions using an infrared laser utilized for sampling.56,57) Since the ions are mostly created by protonation/deprotonation processes, the ionization mechanism is extremely similar to that of ESI or DESI.58) Due to two important characteristics, IR-MALDESI can be used for high-throughput analyte detection. First, because exogenous ice is the only matrix utilized during studies, IR-MALDESI ablates and ionizes analyte in a single coordinated process. Second, IR-MALDESI requires few to no sample preparation steps.59)
A standardized workflow for a typical in situ analysis of N-glycans by MALDI-MSI is shown in Fig. 3.
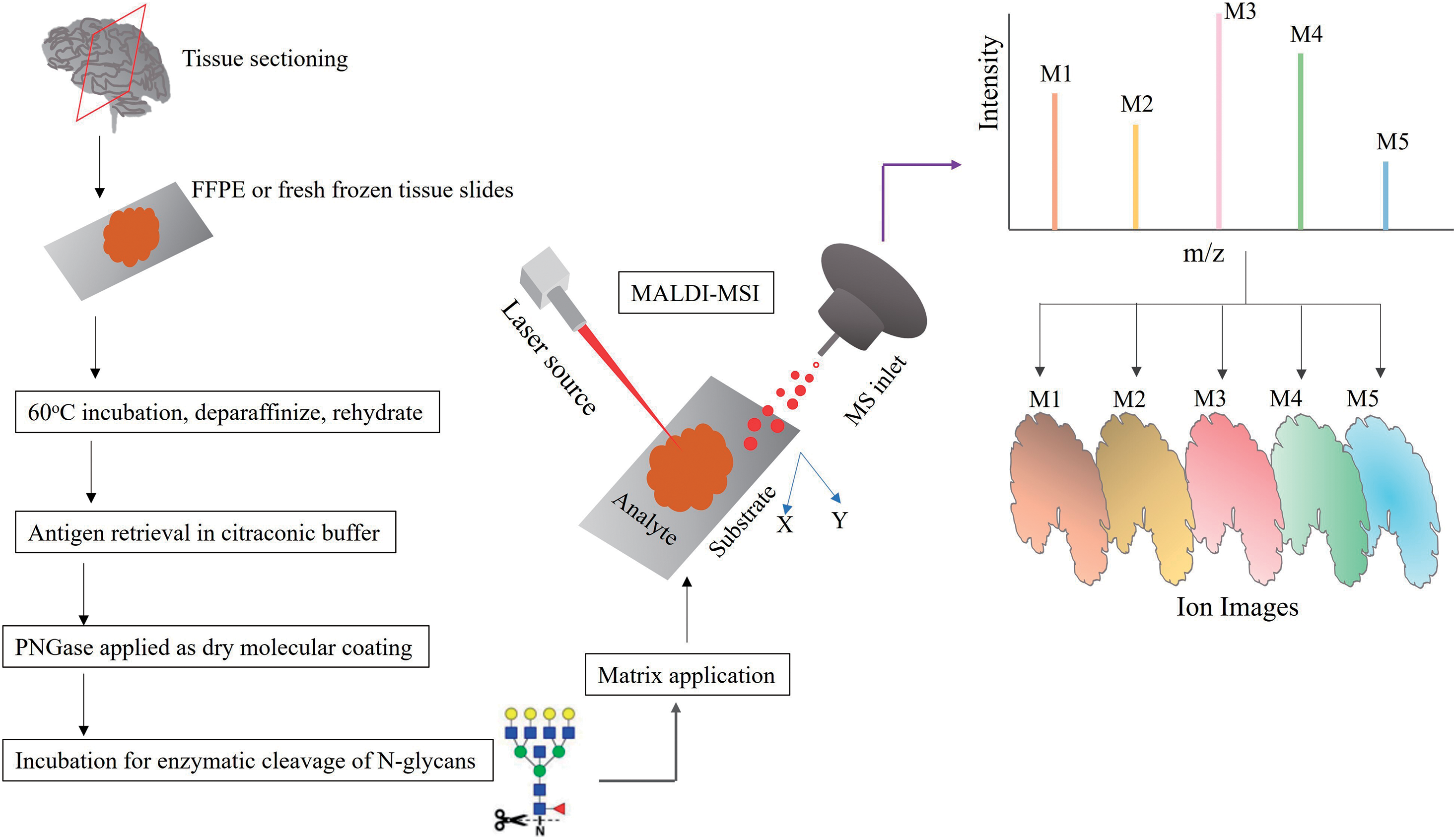
A typical MALDI-MSI workflow includes a) preparation of formalin-fixed, paraffin-embedded (FFPE) or fresh frozen (FF) slides, b) dewaxing and rehydration of tissue specimen, c) antigen retrieval in citraconic buffer, d) PNGase F applied as dry molecular coating, e) incubation period to enable enzymatic cleavage of N-glycans, f) matrix application, and g) MALDI-TOF or MALDI-FT-ICR imaging.60)
Different sample preparation stages must be completed depending on the molecular class and kind of tissue of interest to be scanned, as shown in Fig. 4.61,62) The steps involved in the sample preparation steps are explained in detail in subsequent passages. The type of tissue to be studied determines the complexity of sample preparation.63) While FFPE tissue often requires a multistep process that includes deparaffinization, tissue rehydration, and antigen retrieval, certain applications for FF tissue only require the application of the matrix. The molecular class to be studied may also require extra chemical or enzymatic treatment.63) Matrix application is the final, crucial step in the MALDI-MSI sampling procedure, and it is required regardless of the kind of prior sample preparation that has been done. Every step has the potential to cause degradation or delocalization of the analyte.63) The process of molecules diffusing or migrating across and away from the tissue is known as delocalization. Delocalization’s intensity varies depending on the molecule, sample, and methodology.64–67) To guarantee the efficient extraction and concomitant ionization of the target analyte in MALDI-MSI, the selection of matrix, solvent system, temperature, and matrix application method are all crucial.61,62,68) Additionally, the application method of the matrix, such as wet coating with a nebulizing sprayer or dry coating, influences the obtained matrix crystal size.69,70)
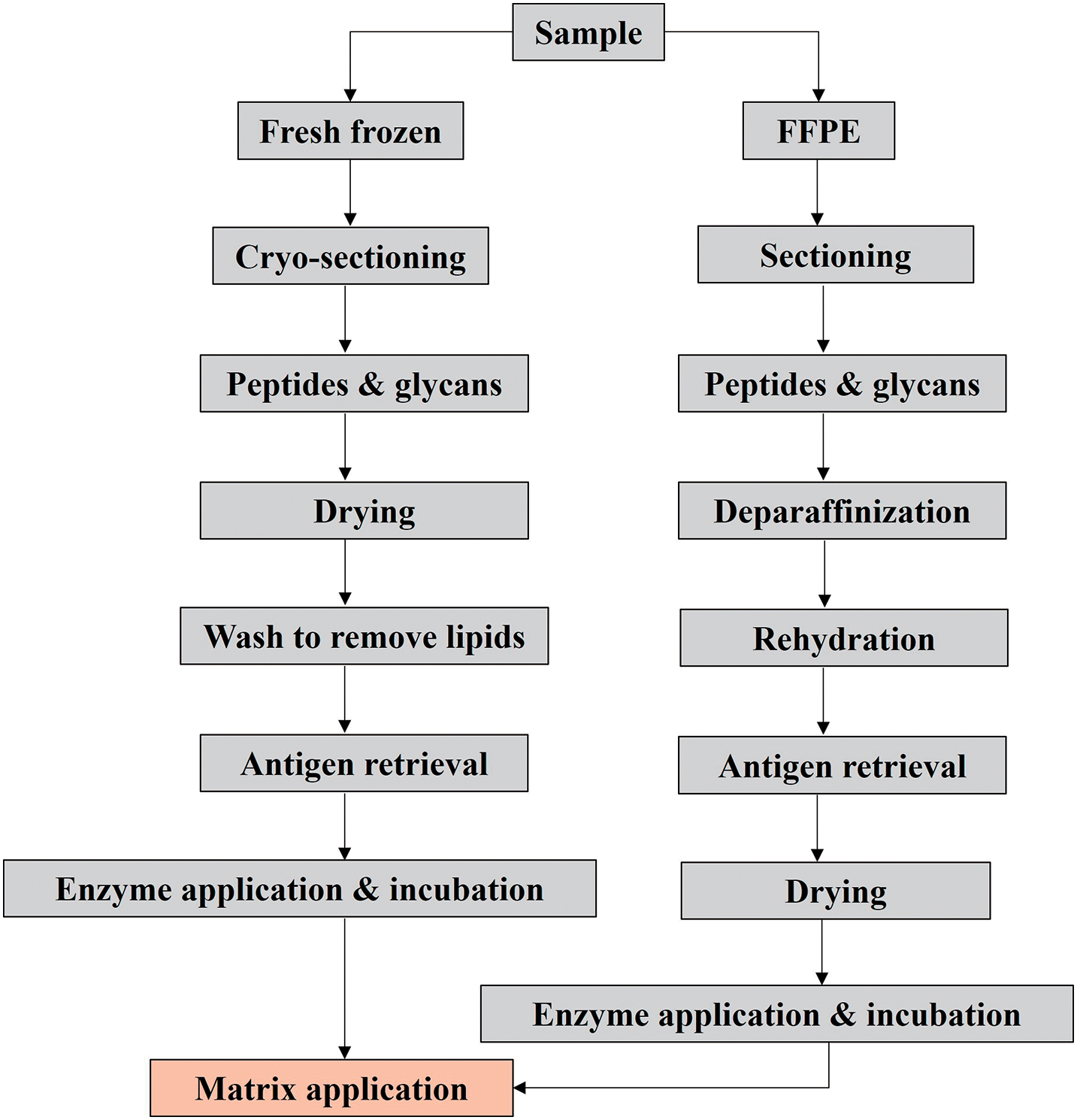
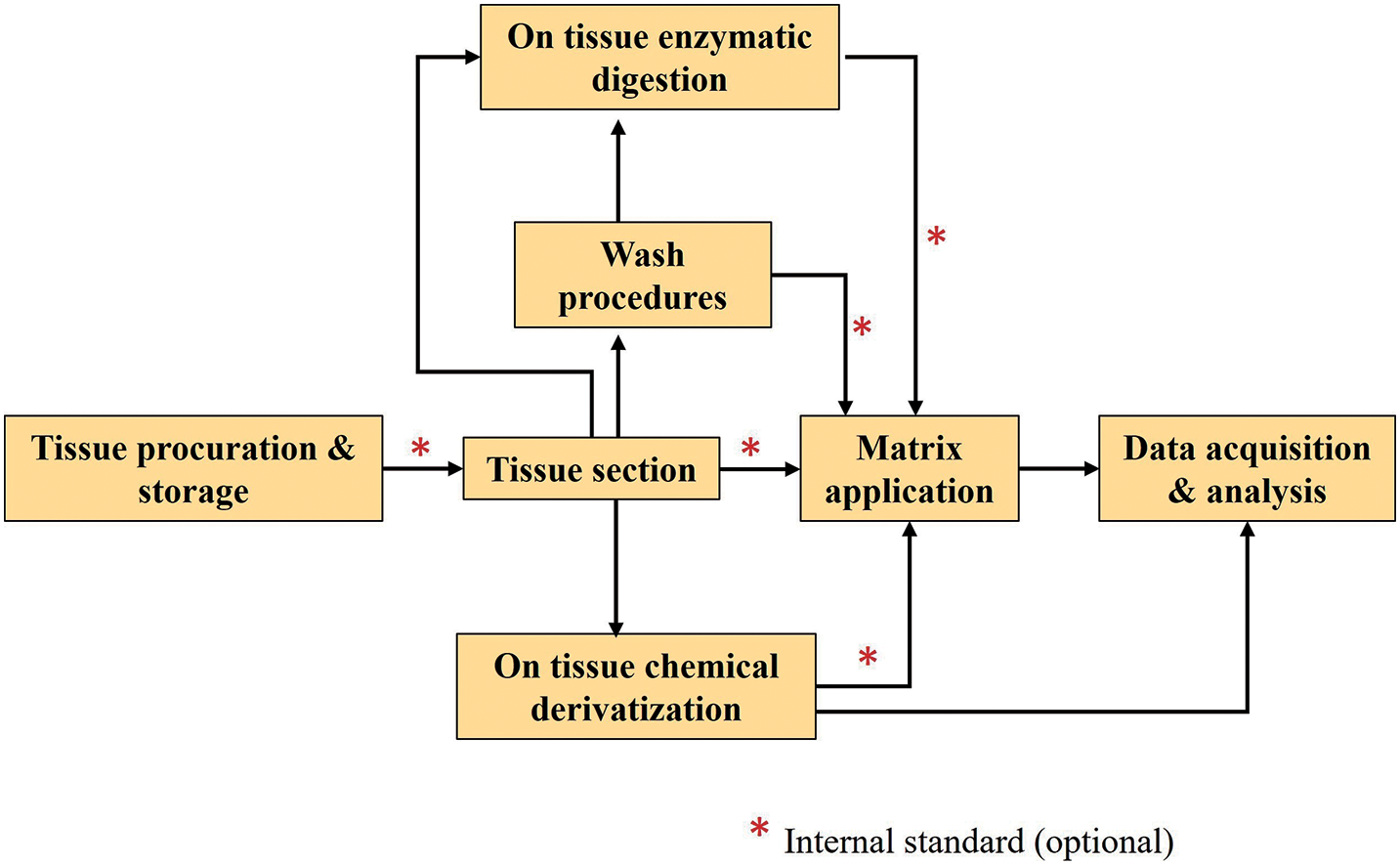
There are multiple components of MSI sample preparation that need to be considered, from sample collection to surface pretreatment ahead of investigation.71) A typical MALDI-MSI workflow highlighting the importance of tissue preparation is shown in Fig. 5. For tissue preparation, many procedures can be used, ranging from tissue harvest to data acquisition.71) Washes, on-tissue enzymatic digestion, on-tissue chemical derivatization (OTCD), and the utilization of internal standard are possibilities in addition to the primary experimental workflow, depending on tissue type, targeted analyte species, and experimental goals.71) Internal standards can be employed for normalization and comparative quantification to counteract the effects of ion suppression in diverse tissue sections. This produces findings that are more highly comparable between different tissues and pixels.72–74)
Tissue mining from animals or human origins and its preservation are the two main variables determining the data integrity for all MSI studies.75) Tissue must be handled carefully and quickly after surgical removal to avoid molecular disintegration and spatial rearrangement.76) Although FFPE and FF tissue are both capable of being evaluated by MSI, FF tissue is the form that is most frequently utilized. The primary reason behind this is that chemical infusion and fixation methods are difficult to incorporate into MS-based studies because they compete with MS detection.76)
The automated and systematic solvent exchange processes necessary to create an FFPE block are likely to enhance and facilitate the systematic detection of N-glycans in FFPE tissue, most likely through the thorough elimination of competing metabolites and lipids.77) Since the majority of N-glycans lack free amino groups, they are unable to crosslink with formalin. For best results, FF tissue also needs to be thoroughly washed to get rid of metabolites and lipids so that the N-glycan signal can be detected.60)
For MSI, the most popular technique for molecular analysis of ex vivo tissue samples are still FF tissue.78) Rapid freezing is necessary to prevent cell wall ripping, which would otherwise alter tissue architecture and the functioning of proteins at the cellular and subcellular levels. As a result, rather than forming a crystalline structure, water solidifies into an amorphous, vitreous state.78) The amount of tissue exposed to the cooling agent is increased when sliced tissues are flash frozen in liquid nitrogen, which theoretically allows for a rapid cooling rate.78) Nevertheless, if a vapor barrier develops on the topmost layer of warm tissue, inconsistent freezing could happen. Isopentane that has been cooled with liquid nitrogen or, rarely, with dry ice is also used for larger samples, notably whole rat brains.78) Desiccated cryosections (5–12 m; 10 m standard) of FF tissues are washed in organic solvents to remove lipids, salts, and other metabolites, therefore enhancing the N-glycan signal. Before enzymatic N-glycan release in FF tissues, antigen retrieval is not necessary, but denaturation by heat enhances the signal. Both FFPE and FF tissue specimens are frequently homogenized for N-glycan extraction along with purification in addition to whole-tissue MSI.79) By doing this, different tissues can be analyzed on a single MALDI target plate, and it can help with fragmentation and further structural confirmation tests that confirm tissue findings.
Thin, flat substrates are essential for MSI analysis. The temperature of cryosectioning, which is frequently used to prepare specimens on a microtome, varies depending on the type of tissue (fatty versus water-based tissues). The dissected tissue slices can be placed simply on a conductive surface for comprehensive tissue imaging. An alternate approach is the sample stretching technique. In this case, the tissue slice is positioned onto parafilm with embedded glass beads and stretched symmetrically in two dimensions. The parafilm is stabilized by being attached to a glass slide. The stretching method improves analyte extraction and spatial resolution.80,81) In either method, the specimen might require to be further prepared after mounting the tissue fragment onto a target.
PNGase F, a peptide N-glycosidase that cleaves between asparagine and the first GlcNAc residue on the chitobiose core of the N-glycan, is incubated with the tissue to release N-glycans from their glycoprotein (GP) carriers during an enzymatic digestion.82) The retention of analyte spatial information in N-glycan MSI is made possible by a “dry” molecular layer of PNGase F that restricts the spread of released N-glycans across the tissue. Current techniques for applying this coating with an automated sprayer system are reliable and remarkably repeatable.83) Tissue homogenates can be used for orthogonal structural investigations after PNGase F-mediated deglycosylation, N-glycan purification, and derivatization procedures, followed by matrix incorporation and spotting for MALDI-TOF MS.84)
The matrix deposition process must be consistent, reproducible, sensitive, and easy to use. The dimension of the matrix crystals also affects how much spatial resolution can be achieved. 2,5-Dihydroxy-benzoic acid (DHB), sinapinic acid (SA),85) 2,5-dihydroxy acetophenone,86) and 4-hydroxy-alpha-cyano-cinnaminic acid are examples of frequently utilized matrices. An excellent technique for reliable and reproducible neuropeptide imaging is87) DHB-based MALDI MSI.82) One feature that all matrices have in common is the crystal size, which influences spatial resolution and calls for careful calibration. Some matrices, including DHB, also result in larger crystals than others. The best solvent settings and nebulizer design factors, however, can help control this issue. Dry matrix application methods like sublimation and dry coating can yield extremely tiny crystal sizes.88,89) While this allows MSI to achieve high spatial resolution, there are limitations because these methods frequently have poor analyte extraction efficiency and little scope for protein imaging.
The choice of matrix deposition depends on the kind of application desired. Automated processes are preferred for large samples and high-throughput research. Because they can become the bottleneck for new, high-throughput biomedical studies, matrix deposition techniques are the focus of intensive research. One of the most important elements influencing the possible cellular spatial resolution is matrix crystal size. When applying a matrix, great efforts have been put forth to reduce the variability of crystal size and minimize its size. Depending on the matrix utilized, modern commercial sublimation and spraying methods can produce crystal sizes between 1 and 30 μm, with sublimation typically reaching even lower crystal sizes.69,90,91) The frontiers of improved spatial resolution are still being pushed by new innovations like electrospray deposition and matrix-free electron-directed soft ionization.70,92)
The analyte(s) of interest should be considered when selecting washing techniques. To prevent analytically disruptive ion suppression during peptide or protein evaluation, salts and lipids must be eliminated. As a result, organic solvents are typically present in the washing solutions.93,94) Additionally, because drug solubility causes displacement or even complete loss from the tissue, organic solvents intended for tissue washing are frequently inappropriate for drug component analysis. The pHs of the washing solutions were modified to values where the medication would have minimal solubility.95) Ammonium acetate buffer at pH 10 was used to wash slices of mouse brain tissue that had been treated with one of three pharmacological compounds. This resulted in a considerable increase in signal intensities when compared to the outcomes of using acidic or neutral buffers.95) Typically, washing processes are left out while analyzing lipids. However, using ammonium formate at pH 6.4 or ammonium acetate at pH 6.7, Angel et al. established a washing protocol for the detection of lipids in negative mode that greatly increased the number of detectable analytes together with their signal intensities.96) van Hove et al. devised a technique wherein a fiber-free paper material was pre-wetted in washing solution and positioned on top of the tissue section for 30 to 60 seconds instead of submerging the tissue in the solution. Smaller tissue segments can be precisely targeted during this washing process, if desired.97) The tissue slices should be somewhat dried before proceeding to the matrix application stage. This can be done by placing them in a desiccator for 10 to 30 min(s) or by quickly applying nitrogen gas. The tissue adhesion to the mass spectrometer slides is stabilized through this technique.
It is recommended to perform many washing procedures ahead of protein or peptide matrix deposition.98) When salts that can react with it are taken out, the matrix can uniformly crystallize, which increases the ionization effectiveness of peptides and proteins. Before applying the matrix for neuropeptide MSI, sophisticated multistep rinsing procedures are often used. These involve pH-optimized organic and aqueous washes designed to precipitate proteins and peptides, remove lipids, and wash away ions that may impede the MSI signal.99–101) However, using aggressive cleaning techniques might damage the specimen and remove peptides that are important for analysis. It is recommended to optimize the matrix and washing processes on adjacent slices to establish the best sample processing protocol to use for each sample type.
The complexity and scale of the spectral data generated during MSI experiments make them difficult to interpret and analyze.102) Multivariate analysis techniques can be used to check MSI data sets for accurate classification.103,104) Multivariate statistics are used to identify the predominant variation and co-variation, which are then included in the appropriate component of the assessment. Essentially, with multivariate analysis, each individual pixel of the spectra acts as an independent sample in a multivariate space. The pertinent loading figures are used to extract the factors, or mass peaks and their scale, that are most crucial for capturing the component’s change.
Principal component analysis (PCA) is a prominent multivariate data analysis technique that is relatively straightforward to apply with MSI datasets.105,106) PCA is used as an autonomous technique to separate sets of variables with significant correlations, such as mass and spatial coordinates. Contrary to PCA, which could contain nonnegative components, probabilistic latent semantic analysis, also known as digital staining, breaks down imaging data into its nonnegative constituents after applying noise reduction and a computerized tissue categorization approach.107,108) PCA and canonical correlation analysis (CC) can be used to correlate MSI data obtained in a variety of ways. The method yields more confined results by correlating the spatial and spectral components of each data set.
Software tools for manufacturer neutral MSI processing have recently become available. To analyze imzML data, the common MSI file format and open-source programs MATLAB, R, and C++ programming have been employed.109–112) Even vendor-specific software can support this open format, which enables the integration of several MSI datasets from various systems. These software applications’ complex analyses are essential for determining how a specific N-glycan’s spatial distribution relates to pathology. The co-localization of key analytes with different tissue morphologies can be visualized by superimposing high-resolution, annotated images of histological staining with N-glycan MSI data.113) Statistical considerations including segmentation, classification, and multivariate analysis are also supported by these programs.114,115) Additional applications involve combining N-glycan MSI data with orthogonal imaging modalities such as IHC and immunofluorescence microscopy, magnetic resonance imaging, and autofluorescence to link glycosylation variations to specific pathological features or biomarkers of interest.116–119)
Because N-glycan imaging is relatively new compared to peptide, lipid, and small molecule imaging, which are more established, there were initially fewer software tools that could facilitate peak assignment.120) Many laboratories currently use shared online databases like GlyConnect or GlyTouCan, or an internal N-glycan database created using software like GlycoWorkbench or GlycoMod.121–124) Software for evaluating N-glycans from MSI-generated spectra is currently in its infancy. Novel applications like Bruker Daltonic’s MetaboScape have shown promise for aligning mass spectra peaks with a large database of known N-glycan masses with matched structures.60)
Prostate cancer, the most prevalent form of cancer in men, is also the second-largest contributor to cancer-related fatalities in men globally.125) Aging, lineage, ethnic origin, food, and way of life are only a few of the variables that affect the onset and progress of prostate cancer.103,126) The “grade group” of the tumor, which is identified by microscopic histopathologic assessment, determines the patient’s outcome in a significant manner.127,128) Although individuals who have low-grade prostate tumors possess a 99% chance of surviving for five years, people with higher grade tumors as well as those who initially have distant metastases have much lower chances of surviving.127) A crucial initial phase in prostate cancer investigation is to recognize the biological characteristics dividing different populations. This may contribute to foundational theories about prostate biology, the development of prognostic biomarkers, and individualized treatment. A variety of biological activities are modulated by the co-translational activity of N-linked glycosylation that occurs during protein folding. According to recent glycomics and proteomic investigations, numerous human malignancies, including breast, lung, and prostate, exhibit significant modifications in both the N-glycan composition and glycosyltransferase expression.129) Furthermore, by modifying the function of extracellular matrix proteins and growth factor signaling, abnormal N-glycosylation has been demonstrated to epithelial-to-mesenchymal transition (EMT) and the resultant metastatic proliferation of cancer cells.130) Determining the N-glycome of prostate cancers could offer a better understanding of the molecular processes underlying the growth of prostate cancer and possibly be utilized to uncover novel indicators or viable therapeutic treatments due to the function of N-glycosylation during EMT and metastasis.131)
Recombinant endoglycosidase F3 (Endo F3), an enzyme having a special affinity for severing core-fucosylated N-glycans linked to glycoproteins, was utilized by West et al. in their MALDI-MSI procedure with FFPE tissues.131) Endo F3 cleaves at the protein binding point between the two main GlcNAc residues, as opposed to PNGase F, which cleaves a wider range of substrates.131) While earlier research highlighted the significance of identifying core fucosylation, the current work discusses the clinical significance and technique of outer arm fucosylation in terms of N-linked glycosylation.131) Numerous malignancies have been connected to the fucosylation of N-linked glycans.132) Hepatic carcinoma progression is specifically connected to alterations after the inclusion of core 1,6-linked Fuc.133) Endo F3 functions more effectively and preferentially on core 1,6-linked fucosylated structures than PNGase F. Focus can be placed on core 1,6-linked fucosylated structures by comparing samples taken from patients to those of healthy controls without the disruption and interference of all the additional complex glycans produced by PNGase F.131)
Proximal fluids from the prostate called “expressed prostatic secretions in urine” (EPSu) are frequently employed in screening and predictive tests for prostate cancer.134) Extracellular vesicles (EVs) and GPs released by the prostate gland are prevalent in these fluids, and their capability to identify alterations in their N-glycan makeup as an indicator of a disorder provides possible novel biomarker contenders.134) In addition to being abundant in EVs, the prostatic fluids, EPSu, and urine are sources for numerous continuing non-coding RNA and associated oligonucleotide-targeted diagnostic techniques for prostate cancer and other disorders.135–137) According to Wang et al. and Erdbrügger et al., the advancement of diagnostic methods for EV derived from urine and EPSu is still an active research domain.138,139) Despite the diagnostic benefits of variations in prostate-specific antigen (PSA) and numerous other prostatic glycoproteins’ glycosylation, procedures to effectively describe N-glycans in urine specimen can be time-consuming and necessitate multiple phases of processing, which prevents their widespread clinical application.140,141) For a sizable number of clinical urine specimens, lectin matrices have been successfully utilized to characterize glycan structures, nevertheless this method is unable to ascertain whole glycan components or distinguish between N-linked and O-linked origins.142,143) With the use of MALDI-MSI for screening, Blaschke et al. created a methodology for comprehensively profiling the N-glycan components of EPSu, prostatic fluids, and urine.134) Complementary findings with N-glycans identified and histologically characterized in prostate cancer specimens have also been presented (Fig. 6). The technique can be used with urine, EPsu, prostatic fluids, and urine EPS-derived EVs, and it has successfully identified over 100 N-glycan components.134) To enhance the levels of protein before spotting, more starting fluid could be concentrated if time to prepare is less of an issue.

N-glycan relative intensities were determined as the absolute intensity divided by the combined nclutotal of all the absolute intensities of the N-glycans found in each sample being compared to account for variations in protein concentrations that might cause higher signal intensities and the detection of more low-abundance N-glycan species. Each sample’s existence or lack of an N-glycan is addressed, and comparisons of the extent of N-glycans found in each sample are also examined.134) Depending on their speculated structures, N-glycan structures were assigned to a class or classes. The N-glycan classes were quantified by adding the relative intensities of the several N-glycans that make up each class. To compare the summed relative intensities of the classes, N-glycan profiles were also investigated by classifying each N-glycan into a group based on the presence and/or absence of Man, Fuc, SA, and sulphate.134) The sum of the multiple sodiated species of sialylated and sulfated N-glycans was used to compare individual N-glycan intensities between samples.134)
The workflow’s concentration and solvent/buffer exchange steps can also be applied to other biological fluids with concentrations of protein comparable to urine.134) The technique can also be applied to enzymes other than PNGase F, such as Endo F3, which is unique to core Fucs (West et al.), or to other glycosidases that are exclusive to different N-glycan structural categories.131) The study offers a preliminary N-glycan profiling workflow that can be used with prostatic secretions and urine. Based on the application and assay’s objectives, it also serves as a scalable framework. If preparation time is less of an issue, starting fluid can be concentrated further to raise protein concentrations before spotting. Additionally, other biofluids with protein concentrations like urine, such as saliva, cerebrospinal fluid, and bronchial lavage, can be adapted to the workflow’s concentration and solvent/buffer exchange step.134)
The structural heterogeneity of N-linked glycans makes it difficult to conduct analytical investigations to determine their biological significance. The liability and ionization selectivity of the monosaccharide present extra difficulties for N-linked glycans containing N-acetylneuraminic acid (NeuAc) (SA).144) To adequately explore the biological significance of N-linked glycans across multiple ailments, these constraints necessitate new analytical approaches targeting these unique obstacles. Because SA consists of a carboxylic acid, it has a negative charge, favoring the negative ionization modality.144) The first in situ IR-MALDESI-MSI analysis of N-linked glycans was demonstrated by Pace et al. in 2022.144) After the enzymatic digestion of N-linked glycans, FFPE human prostate tissue was examined in the negative ionization mode. Based on earlier identifications recognized in the literature, an assortment of 53 N-linked glycans were effectively identified.144) SA residues were present in over 60% of these identifications, demonstrating the ability of IR-MALDESI to safeguard these labile molecules (Fig. 7). In an individual scan, GlycoHunter identified a sizable number of multiply charged peaks, which suggested several chlorine adducted molecules as well as possible sulfate and phosphate alterations.144,145) The study demonstrated that N-linked glycan imaging is possible without the need for chemical derivatization.144)

Due to their molecular variability, N-linked glycans can perform a variety of biological functions when they are connected to asparagine sites on the GP framework. According to Kuo et al., glycocalyx is responsive to the temporal physiological condition of the cell and represents intrinsic cell wellness.146) As a result, it has been thoroughly investigated how the shape and make-up of the glycocalyx alter in response to various disorders.147) The glycocalyx controls relationships with the surrounding atmosphere by serving as a support, barrier, filtering system, and effective biochemical or enzymatic mediator.147) A distinguishing feature of the glycocalyx, an extensive structure of glycoconjugates found on the cell surface and the extracellular framework, is the existence of SA carbohydrates on glycoproteins and glycosphingolipids. SAs are recognized as elements in well-known carbohydrate tumor antigens,148,149) influenza virus-binding receptors, along with other transmissible diseases, and they play a variety of biological processes as parts of glycoconjugates.150–152) Various chemical combinations of SAs are found in biological systems, with NeuAc constituting the most prevalent kind in humans.153,154) Glycoproteins include SAs linked in a variety of anomeric connections, primarily as α-2,3 or α-2,6 isomers as well as various α-2,8 and α-2,9 isomers.154) The sialylated glycosphingolipid class known as gangliosides, which is particularly prevalent in the brain, is made up of many sialylated species joined by α-2,3 and α-2,8 connections.155) The elimination of the SA component owing to its unstable quality has traditionally been a significant problem for usage with laser desorption and ESI sources for the investigation of sialylated glycoconjugates by MS.156–158) SAs are being intensively addressed with various chemical transformation approaches due to their significance in biological sciences and the need for better functional investigations in MS techniques.154,156)
Numerous metabolic, chemoenzymatic, or chemical tagging techniques have been used to add bioorthogonal functional units, such as azides and alkynes, to SAs and along with additional sugar equivalents.159–161) By adding alkyne or azide moieties to SAs that are connected at the α-2,3, and α-2,8 positions, Lu et al. created bioorthogonal chemical labelling probes.162) Pancreatic and prostrate FFPE specimens that have previously been identified by N-glycan MSI163,164) were used to further analyze the amidation reactions (Fig. 8).162) Additionally assessed were human serum and pancreatic cell culture that have been previously identified for sugar antigen expression.165,166) The study’s adaptation of the AAXL and AAN3 techniques streamlines the current workflows for amidation of bioorthogonal sugar azides and alkynes167) and N-glycan MSI168,169) to produce novel techniques with distinctive properties. First off, while α-2,6 SAs along with other carboxylate-containing constituents are not amenable to amidation, α-2,3, and α-2,8 SAs coupled to proteins and lipids are amenable to amidation.162) Second, the AAXL and AAN3 reactions can bind an alkyne or azide to SAs devoid of the use of a living organism with a functioning metabolic process or externally supplied glycosyltransferases and substrates.162) Third, the chemical components required for the direct AAXL and AAN3 reactions are inexpensive, easily accessible, and part of well-established chemical labeling procedures.162) With conventional labeling efficiencies of 95% in clinical tissues and biofluids, AAXL/AAN3 requires a mere four hours of sample preparation to be added to a standard N-glycan IMS procedure.162) Overall, the development of the bioorthogonal click chemistry components by AAXL and AAN3 allows for several novel research methodologies for workflows that target labeling, identification, illustration, and glycoproteomic workflows for SA isomers.162)

Hepatocellular carcinoma (HCC), notably, was found to be more closely linked to N-glycosylation alterations than other types of cancer.170–172) HCC is the second most common cancer worldwide, and its prevalence continues to increase in the United States. Initial evaluations of alpha-fetoprotein revealed core fucosylation as a crucial sign of hepatic abnormalities in HCC, which sparked curiosity concerning liver cancer glycosylation.173,174) Hepatoblastoma (HepG2) cells’ N-glycan profiling revealed afucosylated, bi- and tri-antennary configurations in addition to the high Man species Hex9HexNAc2, indicating that distinct liver tumors with diverse origins have varied glycoprofiles.175) A significant portion of the N-glycans containing a minimum of one Fuc residue found in the HCC tissue microarray, particularly those linked to decreased survival, are core-fucosylated, according to complementary evaluation.133)
DelaCourt et al. (2021) analyzed HCC tissue specimens with MALDI-MSI and compared the spatial N-glycosylation against a commonly used HCC molecular taxonomy to ascertain the association between HCC-specific N-glycosylation alterations and genetic tumor traits.176) Given the obvious distinction in fucosylation trends among S1 and S2 cancers, the fucosylation in S2 tumors is of special relevance (Fig. 9).176) Contrary to what can be observed in S1 tumors, there is a lesser amount of fucosylated components in S2 tumors when weighed against nearby non-HCC liver tissue.176) Although the neighboring nontumor liver tissue expresses more fucosylated entities than an S2 tumor does, these structures have been demonstrated to be closely tumor correlated in S1 cancers. Independent of the tissue surrounding the tumor, fucosylated structures that are typically tumor correlated in S1 cancers are expressed at considerably greater concentrations in S1 tumors than in S2 tumors.176) This is demonstrated by an examination of individual glycan structures. The encoding of fucosylated structures is diminished in S2 tumors, but fucosylation seems to be directly implicated in the growth of many S1 malignancies.176) Developing biomarkers for certain HCC subtypes enhances the likelihood that these markers will function in harmony to improve responsiveness to the beginning stages of HCC, thereby constituting the most promising way to lower HCC-related mortality.176)

Cholangiocarcinoma (CCA) is an epithelial cancer that develops in the biliary mucosa lining, the ducts that convey bile between the liver and the small intestine.177) The second most widespread and possibly terminal form of liver cancer after HCC is CCA.178) The anatomic position of a CCA determines whether it is classified as a distal CCA, perihilar CCA, or intrahepatic cholangiocarcinoma (iCCA) subgroup.179) An accurate diagnosis of CCA in its infancy remains difficult despite progress made thus far in identifying potential risk variables and the root causes of the disease. An assortment of diagnostic techniques is required to diagnose CCA, and a biopsy needs to be performed when viable and considered for the final determination.179) The variability of the tumor’s position, dimension, and pathologic and cellular features is not taken into consideration by traditional diagnostic techniques.177,178) A GP’s N-glycan structure can be changed by either including or eliminating N-glycan residues, of which a number has been linked to liver disorders. Nevertheless, little is understood concerning the N-glycan changes connected to iCCA. N-glycosylation changes have shown great potential as an indicator for several cancers, including HCC, which means there is a pressing need for identifying credible predictive biomarkers for individuals with iCCA.180)
The examination of additional disorders, particularly those affecting the liver, must be pursued using N-glycan changes that were associated with tissue and serum assessments.179) Ochoa-Rios et al. used N-glycan MALDI-MSI to determine the changes that occur immediately in blood and tissue samples from iCCA patients (Fig. 10).180) Whole tissue investigations were performed to better comprehend the histopathologic origin of the N-glycan elements in the iCCA and HCC tissues. N-glycan alterations were markedly more abundant in the iCCA tissue and serum samples compared to HCC and bile duct diseases such as primary sclerosing cholangitis.180) The development of an algorithm that could serve as a biomarker for iCCA was based on the N-glycan changes found in iCCA specimens.180) In comparison with carbohydrate antigen, currently considered an acceptable indicator of CCA, the findings of this study demonstrate that the biomarker methodology increased the iCCA detection accuracy. This study explains the N-glycan changes that happen in the iCCA tissue directly and uses that knowledge to identify serum biomarkers that are capable of being utilized to facilitate the noninvasive identification of iCCA.180)

The primary contributory factor to cancer-related fatalities among gynecological cancers in women is OC, which is the sixth most prevalent cancer among women resulting in death.181,182) The high rate of fatality is mostly brought on by insufficient early diagnosis, which is brought on by cloudy late-occurring symptoms and a dearth of precise early diagnostic indicators. As a result, modified protein glycosylation garnered interest as a prospective source of new supplementary diagnostic indicators given the lack of protein-based indicators allowing OC diagnosis at an early, possibly treatable phase.183) Since OC includes a multitude of diverse etiological and histopathological subgroups, it is far from being an isolated illness. Approximately 90% of all incidences of OC include the most prevalent type, epithelial ovarian cancer (EOC). EOC can be divided into two different groups using genomic techniques. Low-grade serous, endometrioid, mucinous, and clear cell carcinomas are prevalent in group I, whereas high-grade serous and endometrioid tumors are prevalent in group II.183) While low-grade OCs, each of which accounts for around 30% of all possible OC cases, are frequently defined by less aggressive characteristics, high-grade serous OC represents the most prevalent and severe of these.184,185)
Since research on protein N-glycosylation has widely documented abnormalities in the OC tumor micro environment, collecting spatial data will provide insight into tumor-specific N-glycan changes in OC growth and advancement.186) To examine N-glycan localization on FFPE OC slices in both early and advanced stage individuals, Briggs et al. used N-glycan MALDI imaging (Fig. 11).186) In comparison with early stage patients, the spatial distribution of oligomannose, complex neutral, bisecting, and sialylated N-glycan groups was restricted to the tumor areas among individuals with advanced OC. These results help us comprehend how the OC disease advances.186) Nevertheless, additional research is necessary to confirm these outcomes with a more extensive tissue microarray patient population and to understand the role of stage-specific glycans in the evolution of OC.186)

Although extensive research on OC-associated N-glycosylation abnormalities at the level of both general and specific serum proteins has been carried out,187–191) less has been learned about changes that occur directly in cancerous ovarian tissues. The most popular method for N-glycosylation profiling is tissue homogenates, which enable more in-depth structural investigations but lack site specificity. Furthermore, it produces data that cannot be immediately connected to tissue structure, resulting in a significant disadvantage in cancer studies.192) To identify tissue type-specific N-glycosylation in more uncommon histotypes of EOC and nonmalignant OC, Grzeski et al. used N-glycan MALDI imaging in conjunction with chemical SA derivatization.192) In addition to high-Man N-glycans, MALDI-MSI analysis showed that EOC tissues are richly embellished with sialylated glycan structures.192) The most significant finding is that the distribution of in situ EOC tissue differs between 2,6- and 2,3-sialylated N-glycan structures, emphasizing the significance of the linkage-specific SA study. Although the data need to be validated with a wider group, they present important information on molecular modifications that are happening within EOC tissue.192)
Colorectal cancerColorectal cancer (CRC) is the third most fatal and the fourth most prevalent cancer to be diagnosed, and both figures have been rising steadily over time.193,194) N-glycosylation has a clear oncological function in CRC.195) N-glycans have been examined in studies using MALDI-TOF MS using CRC tissue homogenate, multiple CRC cell lines, and serum from patients.196,197) These studies found numerous N-glycan structural characteristics that are unique to CRC tumor tissue, such as elevated Man levels, core fucosylation, sialylation, enhanced branching, and even sulfation. However, human blood specimens or cell culture samples were used in research on the glycosylation alterations in CRC. Studies incorporating CRC tissue samples were carried out on homogenized tissue, so little morphological detail indicative of molecular intratumor diversity was retained.198)
Boyaval et al. published the findings of N-glycan MALDI-MSI following linkage-specific SA modification on a population of patients with stage II CRC with the objective of detecting glycomic intratumor variability while offering novel molecular understanding that might facilitate precise patient differentiation.199) Sialylation and high-Man glycan concentrations tend to be greater in cancer cells than in healthy epithelial cells, although fucosylation and highly branched N-glycan concentrations were lower in cancer cells.199) It suggests that the cancer N-glycosylation signature penetrates the neighboring stroma at the invasive front of the tumor when examining the contact between the cancer site and its surroundings.199) Among this sample group, the finding seemed more pronounced in patients who had poorer outcomes. The results show that N-glycan MALDI-MSI can be beneficial for tissue characterization with possible utilization in molecular pathology; nevertheless, they are still awaiting confirmation of the study’s findings in additional research with larger CRC populations.199) These glycosylation alterations may one day be used to better categorize those suffering from stage II CRC.199)
Local removal of early CRC may be curative, but it is challenging to detect early lesions with a high chance of developing regional metastases, as this greatly influences the types of treatments available.178,199) Numerous cancer types, including CRC, have been linked to both the onset and advancement of variations in sugar moieties.200) Histological criteria-based T1 CRC staging is still demanding, and pathologists’ grading of these criteria varies significantly. It is essential to understand the biology underlying the development of an adenoma into a T1 CRC. Glycomic studies have been published extensively on changes to the N-glycomic pattern in CRC; hence, examining these changes may provide new information about how T1 CRC evolves.200)
To spatially characterize the N-glycan species in a group of pT1 CRC specimens, Boyaval et al. used MALDI-MSI.200) The detected N-glycans’ structural details were ascertained using capillary electrophoresis (CE)-ESI-MS/MS. Comparative intensities and glycosylation properties were determined using an array of 58 N-glycans.200) Analysis revealed distinct variations between tumor sections, dysplastic, and normal epithelium. In the dysplastic section compared to the cancer section, there were more high-Man-type N-glycans, which is consistent with enhanced cell proliferation.200) Researchers noticed alterations in the cancer’s invasive front, including a greater expression of 2,3-linked SAs that matched the region’s glycosylation pattern. The pathological definition of cancer stages may be aided by this study’s ability to distinguish between pre-tumors and tumors that are malignant.200)
Seizures and dementia are examples of neuropathological symptoms associated with congenital glycosylation abnormalities.201) Changes in N-glycosylation will eventually disrupt the machinery in cells, which will affect the functions of glycoproteins.201) Consequently, studying N-glycosylation modifications in a neuroinflammatory context may present an avenue for prospective therapies.201) Although less is known about N-glycans’ participation in brain physiology, it is known that they are crucial for neuronal differentiation and advancement, synaptic growth, and myelinogenesis.202) In order to comprehend how glycosylation is possibly influencing the etiology of neurological conditions, it is crucial to completely characterize the brain’s N-glycome, especially in neurological conditions like neuroinflammation.201)
In an illustration of lipopolysaccharide (LPS)-induced neurological inflammation, Rebelo et al. investigated the function of N-glycosylation.201) LPS, a potent immunostimulant found in the cell membrane of gram-negative bacteria, principally binds to the many TLR-4 receptors present in microglia,203,204) in addition, a lesser degree, in neurons and astrocytes.205–207) The stimulation of transcription factors by TLR-4 along with consequent pathways in microglia leads to the activation of the aforementioned detrimental inflammatory chemicals.208,209) Sialylation and fucosylation significantly decreased (7.5% and 8.5%, respectively) after LPS administration, suggesting that such downregulation may be a key potentiator of inflammatory pathways. Their function in this phenomenon may also be corroborated by an overall increase in oligomannose (13.5%) and the presence of bisected N-glycan structures in the damaged tissue.201) Numerous glyco-analytical techniques have the potential to be leveraged to effectively understand the brain’s glycomic makeup, opening up new research possibilities.201) Tracking fluctuations in drug concentrations during an investigation and their final concentration close to the site on which they function has long been a problem in the process of developing new chemical probes and medications. Dosage assessment is a further objective for assessing the therapeutic potential in terms of safety and effectiveness.210,211) Drugs that target specific cell sections may help improve medication action and absorption as well as transport systems. Another important factor is how medications are distributed within cells.212–214) In order to quantify substances within cells with high resolution and accuracy, it is important to develop and improve diagnostic methods.215–217)
It has been established that N-glycans with negatively charged groups, such as SA, phosphate, or sulfate groups, have significant physiologic effects on the central nervous system.218–220) Particularly, SAs have been demonstrated to have a crucial role in immunological control as well as nervous system growth, plasticity, and repair.221)
The inaugural use of IR-MALDESI-MSI toward the identification of brain N-linked glycans was reported.222) The study further offered a comparative investigation of the effect of tissue segment thickness on tissue glycan imaging and identification.222) The study found that there were roughly 136 N-glycans in the mouse brain due to the increased sensitivity of IR-MALDESI toward N-glycan identification.222) SA residues were present in 50% of the observed N-glycans, which is around three times more than in earlier publications.222) It has been determined that sulfated N- and O-glycan epitopes play an integral part in a variety of biological recognition processes and pathophysiological repercussions.223–225) Nevertheless, the discovery of these low-abundance yet significant sulfated glycans has resulted in the establishment of novel enrichment tactics and approaches for sulfoglycomics.226,227) The research study also published the preliminary glycan MSI analysis to document the N-sulfoglycans in the analysis of brain tissue (Fig. 12).222) Additionally, the ambient, in situ analytical settings avoid the requirement for chemical modifications to identify and stabilize the sulfated glycans. Sulfated glycoproteins were discovered to be multicharged species. It is noteworthy to point out that the corpus callosum, which is distinguished by the occurrence of a cluster of myelinated nerve fibers, has a greater concentration of these sulfated N-glycans than other regions.228) It has been established that sulfated N-glycans have a role in the myelination of the peripheral nervous system.229) Axonal dysfunction caused by demyelinated axons promotes neurological impairment that is irreversible. Myelination is involved in tuning axonal activities.230,231) The spatial distribution of the sulfated glycans in the brain’s myelin-rich corpus callosum highlights the importance of spatial glycan signals in disease pathophysiology in several myelin related illnesses. In conclusion, IR-MALDESI-MSI offers a highly sensitive platform for glycan detection for identifying disease- and/or tissue-specific glycosignature in the brain while maintaining sialoglycans with no chemical derivatization.222)

MSI has been shown to be an efficient technique for examining the in situ spatial localization of molecular species. In contrast to other, generally accepted biochemical imaging approaches, MSI enables label-free screening without requiring any prior understanding of the prospective target species. The sample preparation can be customized for several categories of desired molecules of interest, even though MSI studies are by default untargeted. The development of MSI as a technique for N-glycan analysis has dramatically increased the scope of glycomics studies pertaining to analyses of specific organ, tissue, and cell types. Understanding the complex roles that different N-glycans play in various diseases has been made possible by documenting the in situ spatial element that makes up an individual N-glycan’s expression.
The approach’s broad applicability implies that, provided suitable clinical specimens are available, every kind of cancer along with noncancer illnesses may be studied. The N-glycomes of many organisms and human organs are still mainly unknown. Although many organisms share the basic structural characteristics of high Man and bi-antennary glycans, several species-specific glycans have been identified that are produced by a larger class of glycosyltransferases and glycosidases than those found in humans.
The structural complexity of N-glycans cannot be fully characterized by a single MS technique. However, N-glycan MSI techniques have been shown to be exceedingly efficient as well as beneficial in terms of offering insight on N-glycan co-localization with tissue histopathology. With advancements in instrumentation, data processing, and the incorporation of other techniques, N-glycan MSI’s ability to differentiate isomers, novel glycans, and complex structures continues to grow. This involves using hybrid instruments, further integrating ion mobility separation, and other ionization techniques, particularly ambient pressure techniques like DESI and IR-MALDESI.232–239) MSI research has contributed to understanding the biological etiology and clinical characteristics of various carcinomas by directly studying metabolic processes in tissue samples. MSI’s continuous development and wider applications promise to shed more light on the molecular mechanisms underlying illnesses and ailments in relation to glycosylation. In the years to come, clinical research facilities may soon combine glycan-MSI systems for disease monitoring and early detection through the identification of specific glycosyl molecular fingerprints.
There is a lot of room for application with the analysis of non-mammalian N-linked glycosylation by N-glycan MSI. Glycan MSI was initially created for N-glycans, but it can be adjusted to a variety of enzymatic and chemical release agents. This means that in addition to N-linked structures, other species that can be detected include O-linked glycans, glycosaminoglycans, and glycolipids.240–244) MSI has been shown to be a unique tool for connecting glucosamine metabolism and storage to the manufacture of N-linked glycans. This was demonstrated in a recent study that included isoamylase into the N-glycan MSI method to liberate glycogen polymers for detection.245) The range of carbohydrate analytes that can be detected by MSI will increase as more specialized enzymes are developed.
MSI can be improved as a comprehensive biochemical visualization system in the areas listed below.
Differentiating isobaric ionsBecause of the in situ identification approach, MSI has issues identifying isobaric ions. Ions with the same chemical makeup cannot be distinguished from one another. The “IMS” technique offers a workable way to get around this restriction. Ion detection is yet another hurdle the MSI approach encounters. Ion characterization can be improved by chemometrics based on a range of complementing components, including isotope abundance and highlighted segments.246–248) Online wet experiments offer additional chemical characteristics, including hydrogen–deuterium exchange during ionization in the liquid form or the post-ionization process in the gaseous phase, that help differentiate isobaric ions as well as the post-acquisition dry approach.249,250)
SensitivityTo acquire biochemical fingerprints, particularly from a submicron-sized substance, without sustaining a significant sensitivity loss, it is required to develop an improved sensitivity in situ benign ionization method. It is important to keep advancing on the creation of novel nanomaterials for analyte attachment, painstaking secondary ionization, complex ion focusing lens platforms, and sample-friendly preparation procedures. This provides a huge area of study that could boost progress in MSI research. As technology develops, the discipline of cancer studies will also advance.
Artificial intelligence and omics databaseMachine learning has been successfully incorporated for understanding the chemical characteristic sequence and direct, precise spatial recognition.251) It is anticipated that more complex data science methodologies and computational techniques will be used with the goal of making the combination of spatial data from different glycomics research investigations easier. For MSI researchers to work together and conduct investigations without limitations, an open-access MSI information repository must be established.252) A spatial omics library with open access is also expected to include biochemical phenotypic atlases from cancer cells as well as essential organs. Unquestionably, the wider MSI and omics communities must collaborate and reach an agreement on this issue.
Data acquisitionFully automated robotic machinery is anticipated to result in a less complex and more reliable system for the automatic handling of tissue samples, expedited MSI data collection, and performance verification. Substantial strides have already been made toward advancing this aspect of commercial design.253) The current standard method for collecting MSI results utilizes a microprobe-oriented spot-by-spot or one line of data at a time mode, which may take many hours or even an entire day to complete. This method is used for larger samples or when better image resolution is required. The MSI will operate more efficiently with quick, brief snapshot-mode data collection.254) This necessitates the use of a microarrayed ion detection method and an intricate MS setup.
Potential clinical applications of N-glycan MSI dataA long-missing spatial element in our comprehension of the contribution of N-glycans in tumor growth and function is filled in by the ability to profile and identify various N-glycan species in the tumor microenvironment in FFPE tissue samples. Targeting N-glycans is an emerging technique in the realm of imaging MS. N-glycan imaging of FFPE samples has opened various clinical diagnostic avenues since the panels of N-glycans acquired can be utilized to create algorithms for classification. Using the rapifleX MALDI Tissuetyper TOF instrument (from Bruker Daltonics) is one of the more straightforward options for a new MALDI-MSI platform.255) The goal of this system’s design was to greatly reduce—by at least five times—the amount of time needed to gather and evaluate tissue imaging data. Furthermore, while keeping the quick analysis times, the laser spot size and raster distances can be regularly measured at 10–20 micron resolutions. Using this novel instrument, the pilot MALDI-MSI of N-glycans from FFPE tissues turned out successful. With these shortened acquisition times, MALDI IMS analysis of standard pathology samples and biopsy tissues obtained during surgery may now be completed in a timely manner suitable for clinical laboratory tests.
Understanding the locations of glycan structures in different tissues makes it easier to create lectin-based tests for direct lectin histochemistry tissue evaluations. Anti-carbohydrate epitope antibodies are also an option; however, they are less common.256) In clinical pathology laboratories, fluorescence or light microscopy is commonly employed to visualize cancer biomarkers in tissue. The use of lectins or antibodies may have a drawback in that they identify glycan structural motifs and classes that may also be found on glycosphingolipids or O-glycosylated proteins.256) The ability to identify N-glycan glycoproteins with specificity may be compromised. It is essentially possible to overcome the limits of the existing MSI techniques for N-glycans, and further advancements in methodology and technology, together with an emphasis on imaging other glycan classes, will contribute to the continued development of this approach.256)
In conclusion, the utilization of glycan MSI techniques has contributed to our comprehension of the glycobiome and the intricate biology and chemistry that control the production, operation, and physiological consequences of these carbohydrates. This technology’s ongoing development and extended applications hold the promise of providing new understandings into the biochemical bases of both health and disease in relation to glycosylation. In the near future, clinical laboratories may incorporate glycan MSI systems for the purpose of early illness detection and surveillance through the identification of certain glycosyl molecular signatures. Clinical pathology can be completely transformed by the introduction of MSI, given its present capacity to increase diagnostic and prognostic skills. The complete potential of MSI remains unrealized due to the pressing need for improvements in consistency in sampling, data handling, and some instrumentation restrictions.
The research was not supported by outside organizations.
There are no conflicts to declare.
The study did not involve human or animal subjects.
Mass Spectrom (Tokyo) 2024; 13(1): A0142