2024 Volume 13 Issue 1 Pages A0150
2024 Volume 13 Issue 1 Pages A0150
We developed a rapid, accurate, and quantitative method for analyzing glucosinolates (GSLs) by combining column-free liquid chromatography (LC) with direct-infusion mass spectrometry (MS). Conventional methods for analyzing GSLs take a long time (20–50 min per sample) to perform compound separation on an LC column. We achieved a shortened analysis time of 30 seconds per sample using a direct-infusion method. Samples were continuously injected by a pump and autosampler on an LC system directly into the MS. Orbitrap MS detected 11 types of GSLs in the extracts of turnip hypocotyls. The calibration curve of a GSL standard showed a linear response over a 6-digit concentration range from 1 nM to 1 mM. In addition, no decrease in the detected intensity of GSL ions in 100 continuous analyses of turnip extracts was observed. This method may be applied for rapid analysis of GSLs and other health-functional or bioactive compounds.
Recent advances in informatics have made it easier to analyze many datasets and produce highly accurate research results in large-scale analyses such as cohort studies. To obtain datasets, many samples need to be analyzed; thus, rapid extraction, preprocessing, and instrumental analysis are required. Recently, various methods aiming to reduce analysis time in agricultural and food analysis, such as selecting superior varieties and ensuring food safety, have been tested.1–3) For example, the rapid and non-invasive analysis of biochemical components, such as lignin, in sorghum grain using Fourier-transform near-infrared spectroscopy has made it possible to quickly evaluate the quality of sorghum as food, feed, and fuel.4) In addition, a rapid detection method for herbicides in fruit juices was developed using evaporative dispersive liquid–liquid microextraction with solidification of suspended organic droplets.5)
Glucosinolates (GSLs) are sulfur-containing secondary metabolites found in cruciferous plants.6,7) Approximately 200 types of GSLs are known to exist and are classified into three types (aliphatic, aromatic, and indole) based on differences in the side chains derived from amino acids.8) When plant tissues are damaged, GSLs are hydrolyzed by the endogenous enzyme myrosinase and degraded to isothiocyanates (ITCs).9,10) ITCs are physiologically active substances that have been reported to have cancer-preventive activity in addition to antibacterial activity.11–13) The content and composition of their precursor GSLs are important indicators for evaluating the health functionality of cruciferous plants, including turnip (Brassica rapa ssp. rapa). Turnip is a vegetable that is eaten worldwide, and many varieties are grown in a wide range of regions in Japan, making it easy to obtain many varieties. In addition, turnip seeds have an extremely high germination rate and are easy to handle, so turnip was used as a plant sample containing GSLs in this study.
The analysis of GSLs is typically performed using a device that combines liquid chromatography (LC) with a photodiode array (PDA) or a mass spectrometer (MS). However, in many cases, it is necessary to analyze multiple GSLs simultaneously, and the time required for these LC separations is a bottleneck. For example, the analysis of Brassicaceae seeds by LC-PDA took 30 min,14) and the analysis of edible parts of five types of vegetables belonging to Brassica oleracea (broccoli, cauliflower, kohlrabi, white cabbage, and red cabbage) required 33 min of separation time.15) Furthermore, LC-MS analysis took 25 min to analyze the leaves of three Brassica oleracea vegetables (broccoli, cabbage, and kale).16)
LC is used to analyze a wide range of compounds by separating them using different columns and solvents. It can also be coupled to many types of detectors, such as PDA and MS. On the other hand, conventional LC often takes a long time to separate multiple compounds. In studies involving the analysis of numerous samples, such as cohort studies, the lengthy analysis time can become a bottleneck. In recent years, nano-LC technology has sometimes been used to reduce analysis time. In most studies using nano-LC, the targets of analysis are proteins and polymers, and there are only a few examples of nano-LC being used for the analysis of small molecules. Some studies have used nano-LC to separate and analyze small molecules, such as phytosterols, tocopherols, anthocyanins, and phytohormones; however, although high-sensitivity analysis was achieved, the separation took more than 20 min.17–20)
Solid phase extraction (SPE) is a useful tool for purifying and concentrating target compounds that is easy to operate and relatively inexpensive. However, since only 1 to 10 samples can be processed at a time, it takes time and effort to analyze multiple samples. Since SPE is a simple procedure, it has become possible in recent years to automate it, and several manufacturers sell automated SPE instruments, although they are very expensive.21,22)
In this study, we attempted to quantify multiple GSLs in a short time by introducing samples directly into a high-resolution MS without using LC. We also investigated SPE conditions for sample purification, attempted direct infusion using an autosampler, optimized the flow rate, and confirmed the effect of continuous analysis on detection sensitivity.
Benzylglucosinolate (BZG), gluconapin (GNP), and sinigrin (SNG) were purchased from Extrasynthese (Lyon, France). All other reagents, including LC solvents, were purchased from Fujifilm Wako Pure Chemical Industries, Ltd. (Osaka, Japan).
Plant materialsTurnip seeds (Brassica rapa ssp. rapa cv. Kanamachikokabu, Miyamakokabu, Omikabura, and Shogoinoomarukabu) were purchased commercially. These turnip seeds were sterilized by soaking in 80% (v/v) ethanol for 10 min. The seeds were sown on vermiculite or paper towel and then grown in an artificial climate chamber. The temperature was set at 15°C with 16 hr of light and 8 hr of darkness without humidity control. Hypocotyls and plantlets were harvested at 3 months and 4 days after sawing and used as samples, respectively. Leaves were obtained from commercially available turnips.
Extraction of plant samplesFifty milligrams of plant samples was frozen with liquid nitrogen, crushed using a multi-bead shocker (MB1200; Yasui Kikai Corporation, Osaka, Japan), and extracted with 1 mL of 80% (v/v) methanol containing 0.1% (v/v) formic acid and BZG (final concentration was adjusted to 100 µM) as an internal standard. The extract was separated into supernatant and precipitated by centrifugation (10,000×g, 5 min, MX-305; TOMY, Tokyo, Japan). All of the supernatant was used in the next step.
SPEA weak anion exchange SPE column (Oasis WAX 1 cc Cartridge; Waters, Milford, MA, USA) was used to purify GSLs. Column conditioning was performed with methanol and 2% (v/v) formic acid. After loading the extract supernatant, the column was washed with 1 mL of both 2% (v/v) formic acid and methanol. Elution of GSLs was performed three times with 1 mL of 50% (v/v) methanol containing 5% (v/v) ammonia.
LC-MS analysisLC (UltiMate 3000 HPLC system; Thermo Fisher Scientific, Waltham, MA, USA) was used as a solvent pump and autosampler. The solvent used was 50% (v/v) methanol, the injection was 10 µL, and the flow rate was 10–300 µL/min. The LC was connected to Orbitrap MS (Orbitrap Exploris 240; Thermo Fisher Scientific). The MS setting conditions were as follows: ion source type: H-ESI, ion transfer tube temperature: 300°C, vaporizer temperature: 350°C, spray voltage: 2500 V (negative mode), scan range: m/z = 150–1000, and resolution: 240,000. Data analysis was performed using FreeStyle software (ver. 1.8 SP2; Thermo Fisher Scientific).
To detect GSLs using Orbitrap MS, 1 µM BZG and SNG were dissolved in 50% (v/v) methanol as GSL standards and injected directly into the MS using a syringe pump. Each proton desorption ion was detected (Fig. 1). The average detected m/z value of BZG in the five analyses was 408.042259, and the error from the theoretical value (408.042848) was 0.59 mDa. Moreover, for SNG, the error between the average detected value (358.026508) and the theoretical value (358.027198) was very small (0.69 mDa). GSLs were detected with high accuracy using Orbitrap MS.

Extraction and purification of GSLs from turnip leaves was attempted based on the method of Sasaki et al.16) One micromolar of BZG and SNG was dissolved in 80% methanol, including 0.1% formic acid. The same solvent was used for turnip sample extraction. The GSL contents in each fraction (1 mL of column through, wash, the first elution, the second elution, and the third elution) were analyzed by direct infusion Orbitrap MS. As a result, 96.1% and 94.9% of BZG and SNG, respectively, were eluted in the first elution fraction (Supplemental Figure 1A). To confirm that GSLs in turnip samples were also recovered under the conditions of SPE, the turnip extracts were purified. The recovery rates of all detected GSLs from turnip leaf extract were over 99.9% (Supplemental Figure 1B). Based on these results, we performed the following experiments under these solid-phase extraction conditions.
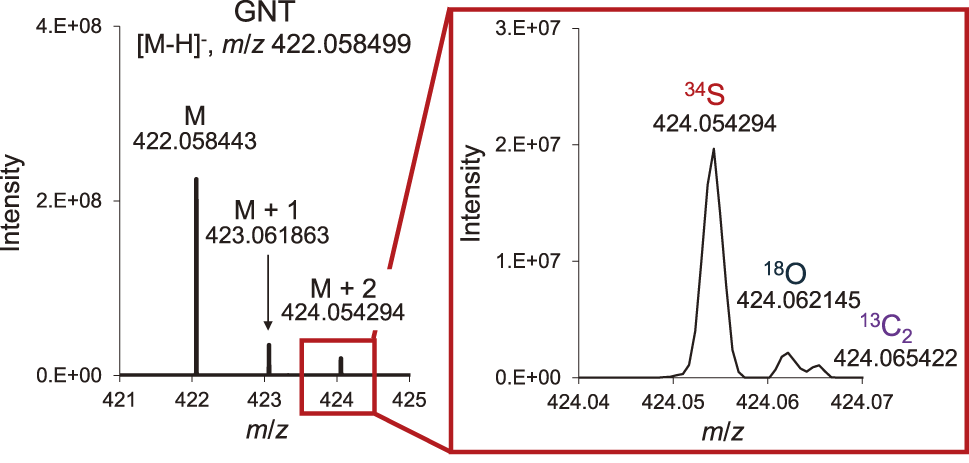
Continuous sample infusion using an autosamplerDirect sample infusion with syringe pumps requires syringe changes for each analysis, which is labor-intensive and time-consuming when analyzing multiple samples. Therefore, we attempted an automated continuous analysis of multiple samples using an LC autosampler. The autosampler outlet and MS inlet were directly connected by stainless steel tubing. First, 10 µL of 1 µM BZG and SNG were injected at a solvent flow rate of 10 µL/min. Peaks of both GSL standards were detected around 2.6 min after starting the analysis (Fig. 2). Next, to further reduce the analysis time, the same GSL standards were injected under higher solvent flow rates (100, 200, and 300 µL/min). In the case of 100 and 200 µL/min flow rates, the GSLs peaks began at 0 min and ended within 1 min and 30 seconds, respectively (Fig. 2). However, in the 300 µL/min condition, the peak area (AA) could not be accurately calculated because the peak had already started to be detected at 0 min. Comparison of AA, peak height (AH), and signal-to-noise ratio (SN) at 10, 100, and 200 µL/min conditions showed that all results were higher at the 200 µL/min condition (Fig. 2). In addition, we attempted to detect GSLs from turnip extract under 200 µL/min conditions that were optimal based on these results. As a result, we detected 11 peaks that were considered to be GSLs. Based on the accurate mass information obtained, these peaks were estimated to be the GSLs that we previously reported from turnip hypocotyls (Table 1).23,24) The differences between the detected and calculated m/z values for each GSL were all less than 0.4 mDa (Table 1). Because GSLs are sulfur-containing compounds, their sulfur isotopes can be detected by high-resolution analysis to distinguish them from isotopes of other elements. In this study, isotopic gluconasturtiin (GNT) peaks derived from 34S, 18O, and 13C2 were identified at masses approximately 2 greater than the [M−H]− peak (Fig. 3). This is strong evidence that the detected peaks from turnip samples were GSLs.

| Classification | Name | Abbr. | Chemical formula | [M−H]− m/z | ||
| Calculated | Detected* | Error** | ||||
| Aliphatic | Gluconapin | GNP | C11H19NO9S2 | 372.042848 | 372.042799 | 0.154 |
| Glucobrassicanapin | GBN | C12H21NO9S2 | 386.058499 | 386.058616 | 0.241 | |
| Progoitrin | PGR | C11H19NO10S2 | 388.037763 | 388.037819 | 0.217 | |
| Glucoerucin | GER | C12H23NO9S3 | 420.046220 | 420.046214 | 0.246 | |
| Glucoberteroin | GBT | C13H24NO9S3 | 434.061870 | 434.061768 | 0.218 | |
| Glucoalyssin | GAL | C13H25NO10S3 | 450.056784 | 450.056787 | 0.188 | |
| Aromatic | Gluconasturtiin | GNT | C15H21NO9S2 | 422.058499 | 422.058335 | 0.238 |
| Indole | Glucobrassicin | GBS | C16H20N2O9S2 | 447.053749 | 447.053746 | 0.213 |
| 4-Hydroxyglucobrassicin | 4HGB | C16H20N2O10S2 | 463.048664 | 463.048842 | 0.345 | |
| 4-Methoxyglucobrassicin | 4MGB | C17H22N2O10S2 | 477.064314 | 477.064354 | 0.261 | |
| Neoglucobrassicin | NGB | C17H22N2O10S2 | 477.064314 | 477.064354 | 0.261 | |
*Detected values are averages of three replicate samples of four turnip varieties.
**The error values between calculated and detected values are shown in mDa.
To clarify the detection limit concentration of GSLs in Orbitrap MS detection and the linearity of concentration-dependent detection peak area values, GNP from 1 pM to 1 mM was analyzed. One micromolar BZG was used as an internal standard. The relative area value was calculated by dividing the obtained peak area value of GNP by the peak area value of the internal standard. We found a slight change in the relative area value from 1 pM to 1 nM; however, an increase in the relative area value that maintained linearity was observed from 1 nM to 1 mM (Fig. 4). This R-squared value also showed a high value of 0.9873. Thus, the detection limit of GSLs in this analytical system was 1 nM, suggesting that concentration-dependent changes in relative area values can be detected in the range from 1 nM to 1 mM. Quantification was performed in subsequent experiments using the approximate curve in this concentration range as a calibration curve.

Among the 11 GSLs contained in the four turnip cultivars, three GSLs (GNP, GBN, and GNT) with peak area values within the range of the calibration curve were quantified. GNT, the most abundant GSL, ranged from 0.32 to 1.25 µmol/g fresh weight (FW). GNP and GBN had lower levels, ranging from 0.05 to 0.29 µmol/g FW. These values were not significantly different from previous reports (Table 2).18) The characteristics of each cultivar were reflected in the differences in GSL content. On the other hand, it is possible that the stress caused by the difference between the growth conditions used in this study and the optimal growth conditions for each cultivar affected the GSL content of each cultivar. The remaining eight GSLs could not be quantified due to their small AAs. Although a 50-mg sample was used in this study, it would be possible to quantify these GSLs present at trace levels in turnips using a larger amount of turnip hypocotyls and concentrating them using the SPE method used in the pretreatment. The results with large standard deviations are also due to inter-sample variation; however, this can be expected to improve as the number of replicates increases. The short analysis time provided by our method is useful for increasing the number of replicates.
| Cultivars | GNP | GBN | GNT |
| Kanamachikokabu | 0.07 ± 0.06 | 0.18 ± 0.13 | 0.32 ± 0.04 |
| Miyamakokabu | 0.10 ± 0.11 | 0.14 ± 0.12 | 0.95 ± 0.43 |
| Omikabura | 0.18 ± 0.07 | 0.29 ± 0.21 | 1.25 ± 0.49 |
| Shogoinoomarukabu | 0.05 ± 0.07 | 0.09 ± 0.07 | 0.78 ± 0.15 |
*Concentration values are averages ± standard deviations of three replicate samples.
In direct-infusion MS (DIMS), which does not involve separation by LC or gas chromatography prior to MS detection, there is concern about matrix effects or ion suppression on quantitative values. The quantitative values of GSLs obtained in this study were not significantly different from those in previous reports, indicating that these effects were limited under the analytical conditions used in our study. Both the use of internal standards and SPE prior to DI would have had a significant impact on the accuracy of the quantification values. Although their high cost is an issue, higher quantification could be expected if stable isotopes of GSLs were used as internal standards. On the other hand, the matrix effects that can occur when using larger amounts of sample may require careful consideration.
Effect on the detected peak area in multi-sample sequential analysesIn our method, samples are introduced directly into the MS after extraction and pretreatment without passing through the LC column; therefore, continuous analysis of multiple samples may result in decreased sensitivity or fluctuating detection values due to the accumulation of contaminants in the MS ion source. To investigate the effect of continuous analysis on the measured values, young turnip plant samples were analyzed 100 times in succession. Figure 5 shows the relative area values of the detected GSLs to the internal standard from the 1st to the 100th analysis. In addition, the relative standard deviation of the measured relative area was low at 2%–6% for all GSLs. These results suggest that continuous analysis of multiple samples is possible using this method.

In this study, we established a rapid analysis method for GSLs using the DIMS method. The time required for analysis by this method was 30 seconds, which was significantly faster than the 25–45 min required by conventional methods. In the analysis of GSLs in turnip hypocotyls, we were able to detect and identify 11 previously reported GSLs through highly accurate and high-resolution analysis. In addition, we were able to obtain quantitative values close to those obtained by conventional methods for the three GSLs mainly found in turnip hypocotyls. In 100 consecutive analyses of GSLs contained in turnip seedlings, small fluctuations in the measured values were observed as the number of analyses increased, indicating that continuous analysis of multiple samples is possible.
Although this study focused on GSLs in extracts of turnip hypocotyls, the rapid analytical method developed can be applied to other compounds. By selecting the SPE column, solvent, and ionization method, it is expected that a wide range of compound groups can be similarly analyzed in 30 seconds. Moreover, because nano-LC allows the separation of compounds using a column, it is possible to separate and quantify compounds that could not be separated in this study using the same molecular formula. Further innovations in nano-LC technology for rapid analysis are necessary. Furthermore, as our laboratory did not have an automated SPE instrument, further studies are needed to assess whether the use of an automated SPE instrument reduces the time and effort required to analyze multiple samples. In addition, robotics can be used to automate the extraction step; thus, robotics combined with automated SPE and DIMS may be able to analyze multiple samples in a shorter time. This method, combined with SPE and DIMS, will be useful for cohort studies, compound screening, and cultivar breeding, which require analysis of multiple samples.
This work was supported by the Tojuro Iijima Foundation for Food Science and Technology.
Supplementary Fig. S1. The recovery rate of GSL standard compounds (A) and GSLs in turnip leaf extract (B) in solid phase extraction fractions. Abbreviations of GSLs are presented in Table 1. CT: Column through, E: Extraction, n.d.: Not detected.
Mass Spectrom (Tokyo) 2024; 13(1): A0150