2014 Volume 3 Issue 1 Pages A0026
2014 Volume 3 Issue 1 Pages A0026
Iron oxide nanoparticle (NP)-coated target plates were employed for the direct detection and analysis of low molecular weight lipids by laser desorption/ionization (LDI) mass spectrometry (MS). We have demonstrated that the use of the iron oxide NP-coated target provides a simple, direct, and rapid detection method for lipid standards and epidermal surface lipids without any cumbersome sample pretreatment as well as mass spectra that are free of background matrix peaks. Lipid standards (1-stearoyl-sn-glycero-3-phosphocholine, 1,2-dioleoyl-sn-glycerol, 1-palmitoyl-2-oleoyl-3-linoleoyl-rac-glycerol, 1,2-distearoyl-sn-glycero-3-phosphocholine) were detected as either protonated or cationated species. Clean MS/MS spectra for each lipid were also successfully obtained. Pre-MS surface cleaning of the target plates with UV-ozone treatment successfully removed organic contaminants that would interfere with the mass spectra especially in the low molecular weight region. Preliminary application of the presented target plate to the detection of endogenous lipids in latent fingerprints showed promising results and for potential use in the visualization and chemical composition determination of latent fingerprints by nanoparticle assistance.
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS), since its development in 1988 by Tanaka et al. has been widely used in various analytical fields for its soft-ionizing and efficient characteristics that allows for the detection of analytes with minimal fragmentation.1) The method took advantage of the inorganic cobalt nanoparticles (NPs) having high surface area-to-volume ratio, which aided in efficient desorption/ionization of analytes. The most commonly used organic matrices for MALDI-MS today include sinapinic acid (SA), 2,5-dihydroxybenzoic acid (DHB), and α-cyano-4-hydroxycinnamic acid (CHCA). Organic matrices used in conventional MALDI require co-crystallization and are well known to have issues with sweet-spot searching and surface inhomogeneity. Ionic liquid matrices such as 3-aminoquinoline/α-cyano-4-hydroxycinnamic acid (3AQ/CHCA) overcome such inhomogeneity issues; however, undesired matrix-derived background issues still pose a problem in the analysis of small molecular weight compounds like lipids. Inorganic matrices including graphene and carbon-based materials have also been explored.2,3) While the use of matrix significantly increases ionization efficiency as well as decreases ion fragmentation with certain matrices, issues still remain with interfering background ions from the matrix itself. For this reason, matrix-free desorption/ionization methods have caught interest in the recent years.
Various surface-assisted laser desorption/ionization (SALDI) methods were also developed using small inorganic particles mixed with glycerol to substitute for the chemical matrix of MALDI since the original report by Sunner et al.4,5) Desorption/ionization on silicon (DIOS)6,7) and other laser desorption/ionization techniques including polymer-assisted (PALDI),8) nano-wire assisted (NALDI),9) and colloidal graphite-assisted (GALDI)10) are other “matrix-free” methods that have been studied previously. A comprehensive review on recent advances in SALDI-MS techniques and their applications can be found elsewhere.11,12)
Metallic and metal oxide nanoparticle assistance is another “matrix-free” LDI-MS approach to detecting and analyzing low molecular weight (<2000 Da) biomolecules. Gold,13) platinum,14) silver,15) ZnO,5) iron,16) TiO2,17,18) and MnO2/MnO319) are only few examples of nanoparticles that have been successfully used for SALDI-MS of small biomolecules such as peptides, fatty acids, lipids, polysaccharides, and oligonucleotides.20) In particular, their chemical stability in air offered an advantage in using metal oxides as SALDI substrates.12) A recent study compared various metal oxide surfaces for matrix-free analysis of various low molecular weight lipids including vegetable oil shortenings and microbial lipids.21) Iron oxide NPs have also been employed previously for the analysis of peptides and proteins by MALDI-MS, although the surfaces of the iron oxide NPs were modified with oleate in the study.22) Other reported examples of iron oxide NPs as affinity probes (affinity-SALDI-MS) include analyzing phosphopeptides using Fe3O4/TiO2 core/shell NPs,23) small-molecule drugs using human serum albumin-modified Fe3O4 magnetic NPs,24) and bacterial endotoxin using chitosan-modified Fe3O4 nanomagnets.25)
In this study, iron oxide NP-coated targets were developed and used for the direct detection and analysis of low molecular weight lipids. One of the disadvantages of metal oxide NPs used in SALDI is in its very own advantage of having a large surface area—the surface can easily be contaminated from air exposure. To overcome the issue of surface contamination that commonly accompanies nanoparticle-coated targets, surface cleaning pretreatment by UV-ozone exposure was applied to each target plate prior to sample analysis. We have demonstrated that the use of the iron oxide NP-coated target provided a simple and rapid detection method for lipids, and also applied this technique to the detection of endogenous lipids in latent fingerprints.
All purchased lipid standards were of the highest purity available. 1-Stearoyl-sn-glycero-3-phosphocholine (18 : 0 LysoPC), 1,2-dioleoyl-sn-glycerol (18 : 1 DG), and 1-palmitoyl-2-oleoyl-3-linoleoyl-rac-glycerol (TG (16 : 0/18 : 1/18 : 2)) were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). 1,2-Distearoyl-sn-glycero-3-phosphocholine (18 : 0 PC) was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). 2,5-Dihydroxy-benzoic acid (DHB) and α-cyano-4-hydroxycinnamic acid (CHCA) were purchased from Shimadzu GLC (Tokyo, Japan). Chloroform, methanol, and acetone solvents were of HPLC grade and were purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). Stainless steel target plate (SUS) was obtained from Shimadzu Corporation. Iron oxide NP-coated target plates were graciously donated by Shimadzu Mass Spectrometry Business Unit (co-manufactured with Dai Nippon Toryo Co., Ltd.).
Preparation of iron oxide NP-coated target plateCommercially available powdered iron oxide with primary particle size of 50–60 nm was dispersed in ethyl alcohol using a typical bead milling process for 12 h. Citrate buffer (aqueous solution of 25 mM citric acid and 37 mM ammonium dihydrogen citrate) adjusted to pH 4.0 was added to the iron oxide dispersion, followed by dilution with aqueous ethanol solution. The resulting product was dropped and equally dispersed onto the sufficiently cleaned SUS target the size of a typical microscope slide using a dispensing device, and then dried under reduced pressure for several minutes (Fig. 1(a)).

Ten milligrams per milliliter stock solutions of the lipid standards were prepared by dissolving the lipids into 70 : 30 chloroform/methanol (v/v) solvent mixtures, and then diluted to 10 μM working solutions. Stock solutions were stored at −20°C in glass amber vials.
Target plate preparationPrior to analysis, SUS and iron oxide NP-coated targets were subjected to UV-ozone surface cleaning treatment with a Photo Surface Processor SSP-110 (SEN LIGHTS Co., Ltd., Osaka, Japan). Plates were exposed to intense UV radiation for two hours to decompose and remove any organic contaminants present on the surface of the target plates.
Mass spectrometryMass spectrometric measurements were performed using MALDI-TOF MS instruments (AXIMA Performance and AXIMA Resonance, Shimadzu/Kratos, UK) in positive ion mode. All spectra were calibrated using a two-point calibration employing coronene (C24H12, m/z 300.3) and fullerene (C60, m/z 720.0). For analyses on SUS target plates using matrices, 0.5 μL of lipid sample and 0.5 μL of matrix were pre-mixed before deposition onto the sample well. Deposited sample-matrix mixture was allowed to air-dry prior to MALDI-MS analysis. For analysis on the iron oxide NP-coated target plate, 0.5 μL of lipid sample was directly deposited onto the sample well and allowed to air-dry.
UV-ozone is a dry surface treatment technology for surface cleaning.26) Removal of organic compounds such as carbohydrates from surfaces by UV radiation is well known. When molecular bonds are applied with energy higher than those of the binding energy, ions, neutral molecules, and free radicals are generated as the molecular bonds are broken. The released organic compounds are oxidized by the UV-generated ozone and are broken down to CO2, H2O, N, O2 molecules and eliminated from the target surfaces. The energy per mol of short wavelength side of UV rays (185 nm) is 472 kJ/mol26); UV irradiation to organic compounds with smaller chemical bond energy will result in its degradation and further elimination from the target surface.
Iron oxide NP-coated target plates were subjected to UV surface treatment for two hours. Microscope and SEM images of a sample well surface of the target before and after UV-ozone treatment are shown in Fig. 1(b). No surface damage or corrosion was observed from the UV-ozone treatment. Conventional surface treatment methods rely on the anchor effect by roughening of the surface which improves the adhesiveness but causes corrosion pitting.26) Treatment by UV is the only surface cleaning method that does not corrode the target surface and therefore is suitable for nanoparticle coated targets such as the one used in this study. Surface contaminants are eliminated to a non-detectable level after two hours of treatment by UV (Fig. 1(c)). After two weeks, some adhesion of contaminants was seen but was minimal and we were able to still obtain clean spectra with negligible amount of background noise.
Mass spectra of lipidsTo demonstrate the mass spectrometric analysis of lipids on the iron oxide NP-coated targets, MS and MS/MS spectra of lipid standards were obtained for our targets and compared with those obtained on SUS targets (matrix-free LDI, MALDI with DHB, and MALDI with CHCA).
18 : 0 LysoPC was detected mainly as [M+H]+ ion (m/z 524.4), with minor peaks also detected for [M+Na]+ (m/z 546.4), [2 M+H]+ (m/z 1047.6), and [2 M+Na]+ (m/z 1069.5) ions, as shown in Fig. 2. High laser power was required for LDI-MS on SUS target in attempt to observe the molecular ion peaks but as a result, unsurprisingly, generated many fragment ion peaks overshadowing the molecular ion peaks of interest. Protonated and sodiated molecule peaks were observed under LDI-MS at very low intensities; instead, fragment ions dominated the spectrum. Fragment ions observed under LDI-MS for 18 : 0 LysoPC include the protonated molecule with a loss of the polar head group at m/z 341 ([M+H−183]+) and the sodiated molecule with a loss of trimethylamine at m/z 487 ([M+Na−59]+). Performance of the iron oxide NP-coated target was comparable to those using SUS with DHB and CHCA matrices in terms of ion intensity and obtained peaks.

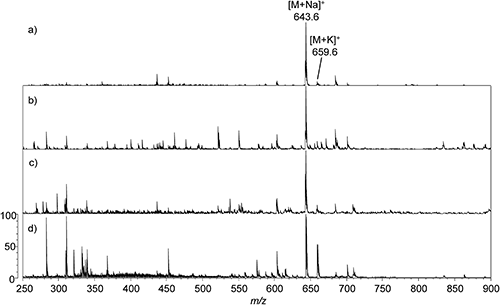
18 : 1 DG was detected predominantly as [M+Na]+ ion (m/z 643.6), although much weaker peak for [M+K]+ ion (m/z 659.6) was also observed (Fig. 3). Iron oxide NP-coated target demonstrated the best performance out of the four compared conditions, producing the cleanest spectra with minimal fragmentation peaks observed. While SUS with CHCA matrix required a lower laser power setting, its spectrum contained abundant noise peaks. SUS with DHB matrix exhibited much more noise peaks compared to the iron oxide NP-coated target as well as peaks deriving from the DHB matrix (not shown).
Molecular ion was not detected under LDI-MS for 18 : 0 PC (Fig. 4(d)). The base peak at m/z 506.4 is derived from the acyl chain. Protonated (m/z 790.4) and sodiated (m/z 812.4) molecules were observed for SUS with DHB and CHCA matrices. The [2 M+H]+ ion peak was also observed for all but the SUS-LDI sample. While the molecular ion peak was the base peak for SUS with CHCA matrix, fragment ion peaks as well as CHCA matrix cluster peaks were abundant producing a noisy spectrum. SUS with DHB matrix and the iron oxide NP-coated target generated similar spectra; however, the molecular ion peak signal was clearly greater using the iron oxide NP-coated target.

As shown in Fig. 5, TG (16 : 0/18 : 1/18 : 2) was detected predominantly as [M+Na]+ (m/z 879.6) and to a much lesser extent, [M+K]+ (m/z 895.6) ions. It is well established that sodium adducts of TGs typically dominate their MALDI mass spectra, and that protonated TGs are normally unseen regardless of the matrix used.27,28) TGs commonly pose problems with MALDI analysis due to prompt fragmentation of the fatty acyl residues that interfere with other TG peaks when analyzing a mixture of TGs (i.e., vegetable oils and biological samples). SUS-LDI required high laser power for the molecular ion to be detectable, and as a result produced a rather noisy spectrum. SUS with CHCA matrix generated many fragment ion and noise peaks that were as strong as or stronger than the molecular ion peak, and at lower m/z range CHCA matrix cluster was also prominent (not shown). SUS with DHB matrix produced a relatively clean spectrum with the sodiated molecular ion as the base peak, but DHB matrix peaks were also observed (not shown). Without having to apply a matrix, the iron oxide NP-coated target produced the cleanest spectra out of the four compared target conditions.

Tandem mass spectrometry was performed on the lipid standards to ensure that MS/MS spectra could be obtained for each lipid with the use of the iron oxide NP-coated targets. Clean MS/MS spectra for each lipid were successfully obtained as shown in Fig. 6. Tandem MS of 18 : 0 LysoPC showed the clear fragment of the polar head which was the base peak of the spectrum (m/z 184) (Fig. 6(a)). The same polar head fragment ion base peak was observed for 18 : 0 PC (Fig. 6(c)), which are typical characteristics of mass spectra obtained from performing tandem mass spectrometry on phosphatidylcholines. Fragment ions of the fatty acyl chains were observed as expected for 18 : 1 DG (Fig. 6(b)) and TG (16 : 0/18 : 1/18 : 2) (Fig. 6(d)). Results confirmed that single and tandem mass spectrometry analyses could successfully be performed on iron oxide NP-coated targets and generated clean spectra with negligible amount of noise peaks.

Thus far we have demonstrated the use of iron oxide NP-coated targets as useful tool in the direct detection of low molecular weight compounds, namely lipids, in this study. As with other surface nanoparticle-assisted LDI-MS approaches, the use of iron oxide NPs for ionization assistance omits the need for matrix use (simple sample preparation) thereby eliminating the issue of matrix-derived interference peaks. Searching for the matrix “sweet spot,” an issue commonly encountered in standard MALDI-MS analysis, can also be circumvented. Larger surface areas available with the use of NPs results in larger analyte loading capacity. Although the approach presented in this paper has been demonstrated to be useful in low molecular weight compounds analysis, macromolecules and other high molecular weight compounds have not yet been tested. In this study, the upper limit of the detected and analyzed compounds is around m/z 1200, and the targets have not been applied for compounds in the higher m/z ranges. Additionally, with standard MALDI-MS, good “matrix-analyte matches” are well established (i.e., CHCA for peptides, DHB for sugars). Such “matrix-to-analyte preference” equivalent has not yet been established with nanoparticle-assisted LDI-MS approach and needs further investigation for the wide array of NPs available. Finally, SALDI-MS is known to have lower sensitivity compared to the results conventionally obtained with organic matrices in MALDI-MS analysis: the sensitivity of SALDI-MS generally lies in the micromolar level. Results obtained using the iron oxide NP-coated targets in this study were comparable, as shown by the S/N ratios at the lowest detectable concentration of the lipid standards used in this study in Table 1.
| Lipid | Lowest detectable concentration (μM) | S/N |
|---|---|---|
| 18 : 0 LysoPC | 1 | 3.0 |
| 18 : 0 DG | 5 | 10.8 |
| 18 : 1 PC | 0.5 | 3.6 |
| TG (16 : 0/18 : 0/18 : 2) | 5 | 10.5 |
Fingerprint samples were collected by pressing the index finger directly onto the iron oxide NP-coated targets. Volunteers were instructed to press their index finger onto the target thrice, holding their finger in place for few seconds each time, to ensure homogeneity between samples as well as to smear the fingermark in order to retain privacy. Figure 7 shows an LDI-MS spectrum of a fingerprint sample taken from a male volunteer. The peaks annotated with “x” are exogenous contaminants that are frequently observed in and have been reported in the past as contaminants arising most likely from common household and personal products.29–31) The contaminants are alkyl benzyl and dialkyl quaternary ammonium compounds that are commonly found in toiletries, personal hygiene and care products, and detergents: dimethylbenzylammonium ion (m/z 304.2), dodecyldimethylammonium ion (m/z 326.6), dimethyldioctadecylammonium ion (m/z 550.6) and its related species (m/z 522.6, 494.6).

Epidermal surface lipids originating mainly from eccrine and sebaceous glands were successfully detected. Detected compounds were identified by analysis of standard reference compounds where possible, and by use of in-house written data extractor to match the m/z values to those of lipids reported in previous studies and registered in the human metabolome database (HMDB, free database http://www.hmdb.ca/search/extractor). The detected lipids included cholesterol [M+H−H2O]+ (m/z 369.4) and protonated free fatty acids (m/z 228.3, 255.3, 257.2, 283.3, 393.3, 382.4) (Fig. 7(a)), LysoPC [M+H]+ and [M+Na]+ ions (m/z 496.6, 518.5, respectively), protonated TG fragments [M−FA+H]+ (m/z 523.4, 551.6) wax ester (m/z 591.5), sodiated DG fragments (m/z 619.6, 640.6), and protonated PC (m/z 732.8) (Fig. 7(b)), and sodiated TGs (Fig. 7(c)). Peaks at m/z 610.5, 638.4, and 664.6 (Fig. 7(b)) are possible lipids but their identities are unknown. Detected compounds are summarized in Table 2. DG and TG ions were detected as sodium adducts as expected due to the high salt contents in human skin surfaces. As mentioned earlier in this paper, protonated TGs are not stable and thus is another reason these species are predominantly detected as sodiated molecules. The TG region spectrum (Fig. 7(c)) is in agreement with other lipid studies using edible oils and human skin surface samples,27,28,32–34) showing characteristic distribution patterns of TGs with 14 Da mass difference reflecting the repeating CH2 units.
| Observed m/z | Compound | Ion |
|---|---|---|
| 228.3 | Myristic acid | [M+H]+ |
| 255.3 | Palmitoleic acid | [M+H]+ |
| 257.2 | Palmitic acid | [M+H]+ |
| 283.3 | Oleic acid | [M+H]+ |
| 309.3 | Eicosadienoic acid | [M+H]+ |
| 369.4 | Cholesterol | [M+H−H2O]+ |
| 382.4 | Tetracosadienoic acid | [M+H]+ |
| 496.6 | LysoPC 16 : 0 | [M+H]+ |
| 518.5 | LysoPC 16 : 0 | [M+Na]+ |
| 523.4 | TG fragment (C30 : 0) | [M−FA+H]+ |
| 551.6 | TG fragment (C32 : 0) | [M−FA+H]+ |
| 591.5 | Wax ester (C40 : 1) | [M+H]+ |
| 610.5 | Possible lipid | |
| 619.6 | DG fragment (18 : 2/18 : 3) | (Na+DG fragment) |
| 638.4 | Possible lipid (DG 18 : 1/18 : 1?)* | |
| 640.6 | Amino-hexadecanoic acid | [M+H]+ |
| 664.6 | Possible lipid | |
| 732.8 | PC 16 : 0/16 : 1 | [M+H]+ |
| 773.6 | TG 14 : 0/14 : 0/16 : 0 | [M+Na]+ |
| 787.6 | TG 14 : 0/15 : 0/16 : 0 | [M+Na]+ |
| 801.6 | TG 14 : 0/16 : 0/16 : 0 or TG 16 : 0/15 : 0/15 : 0 | [M+Na]+ |
| 815.6 | TG 15 : 0/16 : 0/16 : 0 | [M+Na]+ |
| 829.6 | TG 16 : 0/16 : 0/16 : 0 or TG 14 : 0/16 : 0/18 : 0 | [M+Na]+ |
| 843.6 | TG 15 : 0/16 : 0/18 : 0 | [M+Na]+ |
| 857.6 | TG 16 : 0/16 : 0/18 : 0 | [M+Na]+ |
| 872.8 | TG 16 : 0/18 : 3/18 : 3 | [M+Na]+ |
*Observed by Bradhaw et al.24)
Iron oxide nanoparticle-coated targets were used for the direct detection and analysis of low molecular weight lipids. We have demonstrated that the use of the iron oxide nanoparticle-coated target provides a direct, simple, and rapid detection method for lipids without any cumbersome sample pretreatment as well as mass spectra that are free of background matrix peaks. Lipids were detected as either protonated or cationized species. Pre-MS surface cleaning of the targets with UV-ozone treatment allowed for the removal of organic contaminants without corrosion damage of target plates, contributing to cleaner background mass spectra. Preliminary application of the presented target plates to biological samples, namely lipid compounds from skin surface (i.e., latent fingerprints), showed promising results and have potential use in the visualization and chemical composition determination of latent fingerprints by nanoparticle assistance. Further examinations of larger sample sets of latent fingerprints as well as other biological specimens using the iron oxide NP-coated targets are underway to investigate the lipid composition differences within age, gender, and health conditions. Information obtained may be beneficial to the fields of forensic science and medicine, in potential individual determination in crime scenes for the former, and disease (bio)markers through lipid oxidation monitoring (i.e., inflammatory diseases) for the latter. The microscope slide-sized target plates make a good candidate for disposable manufacturing, in view of single patient-use for medical testing. With the use of low-cost disposable target plates, application of this technique can be widened to use in general physical examinations.
This work is supported by the Japan Society for the Promotion of Science (JSPS) through the “Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program).”