2015 Volume 4 Issue 1 Pages A0040
2015 Volume 4 Issue 1 Pages A0040
In vivo concentrations of cellular signaling mediators such as inflammatory mediators are normally maintained at very low levels due to their strong ability to induce a biological response. The production, diffusion, and decomposition of such mediators are spatio-temporally regulated. Therefore, in order to understand biochemical basis of disease progression and develop new therapeutic strategies, it is important to understand the spatiotemporal dynamics of the signaling mediators in vivo, during the progression of disorders, e.g., chronic inflammatory diseases; however, the lack of effective imaging technology has made it difficult to determine their localizations in vivo. Such characterization requires technical breakthroughs, including molecular imaging methods that are sensitive enough to detect low levels of metabolites in the heterogeneous tissue regions in diseased organs. We and other groups have attempted to fill this technical gap by developing highly sensitive imaging mass spectrometry (IMS) technologies. To date, we have established two key techniques toward this goal, including (i) a sample preparation procedure that has eliminated the problem of the postmortem degradation of labile metabolites, and (ii) on-tissue derivatization of metabolites, which can enhance analyte ionization efficiency. Here, we review recent progress in the development of these technologies as well as how the highly sensitive IMS technique has contributed to increasing understanding of the biochemical basis of disease mechanisms, discovery of new diagnostic markers, and development of new therapies.
In molecular biology research, localization of transcripts is commonly visualized with the use of oligonucleotide probe in situ hybridization, and localization of proteins is visualized using immunohistochemistry based on antibodies. However, to date, there is no similar standard technology for the imaging of small metabolite molecules. For example, to determine the biochemical basis of disease progression, identification of the spatiotemporal changes in cell metabolism in vivo is required. Recently, imaging mass spectrometry (IMS; also referred to as mass spectrometry imaging) has attracted great interest for monitoring of cellular metabolism.1,2) The emergence of IMS technology coupled with MALDI,3,4) secondary ion mass spectrometry (SIMS),5,6) desorption electrospray ionization (DESI),7,8) and other desorption/ionization techniques9–11) has added another attractive metabolite imaging technique to the established molecular imaging methods. In addition to its capability for simultaneous visualization of multiple metabolites, there are several other unique advantages of IMS, including its label-free principle, which has led to the emergence of a high number of IMS studies in biological12–15) and medical research16–18) in recent years focused on the analysis of small molecules.
Currently, two IMS applications for analyses of small metabolites, namely, visualization of endogenous small organic compounds19,20) and exogenous compounds such as administrated drugs,21–23) are recognized as practical tools in these fields. Moreover, the former application has attracted particular attention because use of both quantitative (i.e., ESI-MS based metabolomics) and qualitative MS approach (i.e., distribution imaging by IMS) for small metabolites can provide a novel opportunity to analyze how much bioactive metabolites are produced by a specific cellular population within a specific location of a tissue in vivo.24,25)
However, given the enormous diversity of metabolite species and their distinct functional roles, the detection sensitivity and selectivity of IMS should be improved to cover the wide range of metabolites. For example, in vivo concentrations of signal-transduction lipids (e.g., prostaglandins, inflammatory lipid mediators) are kept at very low levels due to their strong ability to induce inflammation. On the other hand, phospholipids, the major building blocks of cellular membranes, are much more abundant within cell and tissues, at levels more than 109 times higher than typical prostaglandins. Although biological tissues are complex mixtures of such metabolite species, the current IMS sample preparation protocols for purifying specific target analytes are limited. Thus, innovation and effort are required to develop novel methods to effectively ionize and selectively detect such trace amounts of molecules that form complex tissue samples.
We and other research groups have approached this problem by developing a highly sensitive IMS method. In particular, we have developed several key techniques, including (i) a sample preparation procedure that eliminates the common effect of postmortem (PM) degradation of labile metabolites,26) and (ii) on-tissue derivatization of metabolites that could enhance the analyte ionization efficiency.27) These techniques have enabled visualization of diverse metabolite species, including not only abundant phospholipids28–30) but also metabolites present in trace amounts such as minor amino acids,31) nucleotides,32,33) neurotransmitters,34–36) and bioactive lipids,37) in various tissues of diseased model animals (as summarized in Fig. 1). We here review recent progress in IMS development, focusing on the two technical improvements mentioned above to achieve visualization of cellular signaling mediators.
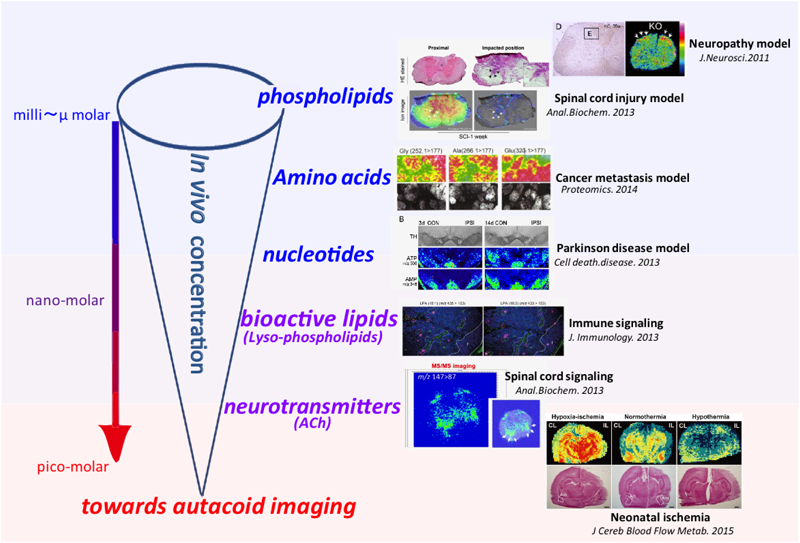
To realize a highly sensitive IMS method, we have focused our initial efforts on accurately measuring metabolites as they are found in their natural state in vivo. However, PM degradation of labile metabolites during the organ sampling process remains a classical problem and critical issue, because major degradation of metabolites tends to occur in tissues within 1 min after death of the animal. Because MS utilizing techniques usually involve sacrificing animals, these postmortem tissues are not suitable for metabolic investigations of organ samples; thus, a key issue to resolve is prevention of the PM enzymatic degradation of metabolites during the IMS sample preparation process. In particular, the brain and heart are especially susceptible to PM changes in the concentrations of labile metabolites.38–40) Moreover, small molecules with high-energy organic phosphates, including nucleotides, as well as signal-transduction molecules such as neurotransmitters, are well known to be sensitive to the PM degradation.25,40) We have approached this problem by rapidly “fixing” the state of metabolites as they exist in vivo. For this purpose, we used a head focused-microwave irradiation method (FMW),41–44) in which the enzymes responsible for the decomposition of metabolites in the brain are inactivated within 1 s.
FMW is essential for analysis of glucose metabolism in vivo by MSIn one study, Sugiura et al.,26) we demonstrated that FMW treatment is an essential process for investigation of glucose-originated metabolic pathways in vivo using MS both for imaging and quantification. The necessity of the FMW treatment arises from the fast and severe PM-induced degradation of brain metabolites. In fact, many classical studies in neurochemistry have described that high-energy phospho-metabolites40) as well as glucose metabolic intermediates38,39) are extremely sensitive to PM degradation. In our study, we showed that the FMW treatment preserved these metabolite concentrations as well as their distribution by halting the effect of PM degradation. As shown in Fig. 2A, when focusing on purine metabolic pathways, we revealed that a conventional decapitation protocol for extraction of the brain caused unacceptable autolytic reductions in ATP and ADP levels, and elevation in AMP levels. Quantification of these adenine nucleotides by capillary electrophoresis-MS showed that 85% of the ATP was degraded into downstream metabolites within 1 min after the decapitation, without FMW treatment. In addition, AMP increased by 10-fold. Importantly, FMW treatment plays a more critical role in IMS than in ESI-MS quantification, because the longer time and additional processes required in IMS measurements could lead to more severe degradation of metabolites. We further demonstrated that the imaging quality of high-energy adenosine nucleotides with FMW was substantially improved compared with that obtained using any other fixation method, with respect to the number of effective pixels and high image contrast (Fig. 2).
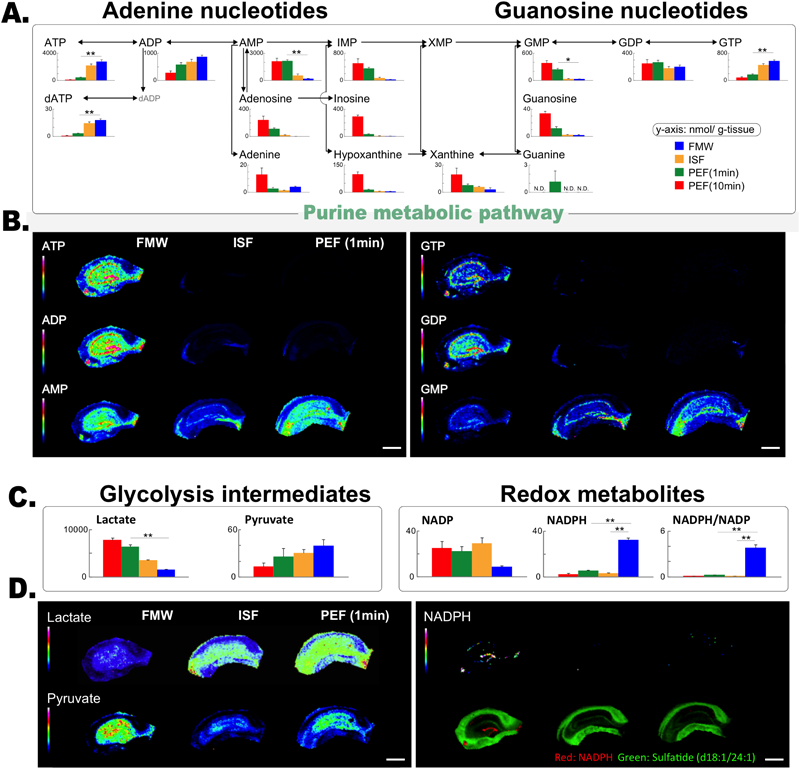
Focusing on purine metabolic pathway, absolute concentrations (nmol/mg tissue) of 15 purine-metabolites (A) and 2 important glycolytic intermediates (lactate and pyruvate, C-left), and a redox-metabolite (NADPH, C-right) were compared among the FMW, in situ freezing (ISF) and conventional decapitation method (posteuthanized freezing; PEF). Significant PM alterations both in quantitative- (A and C) and imaging-analyses (B and D) were suppressed in the FMW-treated sample.
As another advantage, FMW preserved the in vivo concentrations and localizations of important glucose-metabolic intermediates, namely, lactate and pyruvate. After animals die, their respiration and heartbeats stop, and therefore all of their brain cells fall into a state of ischemia. In such conditions, the cells cannot utilize the tricarboxylic cycle pathway because of oxygen depletion, thus enhancing the flow of the glycolytic pathway. Consequently, lactate should accumulate, whereas pyruvate is reduced under this state of PM whole-brain ischemia. As expected, the rapid inhibition of PM enzymatic activity by FMW efficiently inhibited lactate elevation compared with decapitation samples (Figs. 2C, D). Moreover, several redox metabolites, including NADPH, were only detected/visualized from the FMW-treated brain tissues (Figs. 2C, D).
Animal welfareIt is important to note that according to the American Veterinary Medical Association Recommendations (AVMA Guidelines for the Euthanasia of Animals: 2013 Edition),45) high-energy microwave irradiation is a humane method for euthanizing small laboratory rodents. It is also worth noting that unconsciousness is achieved in less than 100 ms with complete loss of brain function in less than 1 s.
The in-situ freezing (ISF) method protected endogenous acetylcholine (ACh) from PM degradation resulting in improved IMS sensitivityThe ISF method, which does not require any specialized instrument, was also proven to be effective for preserving labile metabolites from PM degradation.24,25,46) In contrast to FMW, the ISF procedure halts the PM enzymatic activity by rapid freezing of the brain tissues, while ensuring adequate blood perfusion until the arrival of the freezing front to avoid unnecessary autolysis. This method has been employed for brain imaging of liable energy nucleotides,24,25,46) as well as the neurotransmitter ACh,47,48) since earlier studies showed that the molecular turnover of ACh is quite rapid.49)
In our another study, we showed that the use of ISF was critical for preserving the tissue ACh concentration, and thus could provide high-quality ACh mapping results.48) The distribution maps of the major ACh signature ions (Fig. 3B) clearly shows that the quality of these ACh-derived ion images were improved in the ISF-treated brain, compared to those prepared with a conventional decapitation procedure. Moreover, the increased sensitivity enabled us to perform the ACh imaging at a higher spatial resolution (30-μm scan pitch). Figure 3C shows the expanded ACh distribution image of the hippocampus, revealing a characteristic ACh localization pattern within hippocampal substructures.
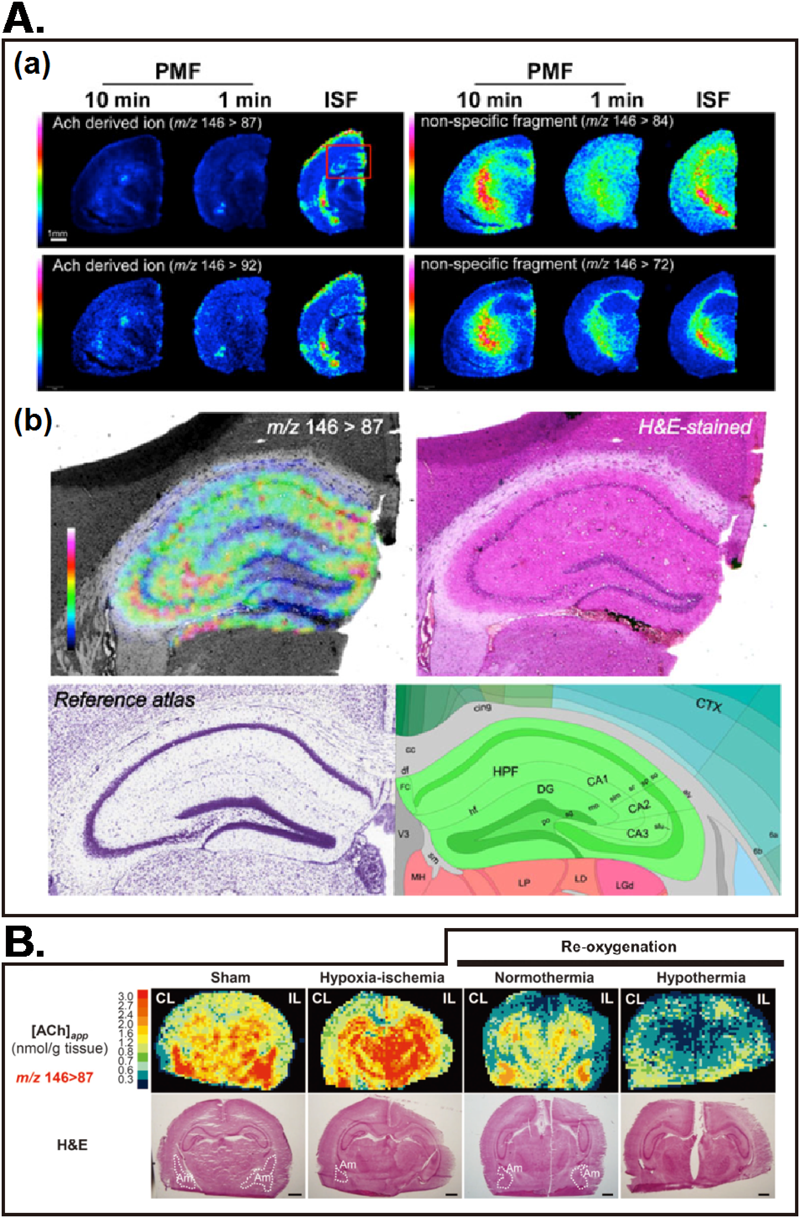
A. (a) ACh signature ion distribution images visualized by MS/MS imaging, obtained from three groups of mice treated with different animal fixation methods; in situ freezing (right) and postmortem freezing with decapitation, in which brain extraction was performed within 1 min (center) and 10 min (left) of decapitation. Imaging results of two major ACh-derived ions (m/z 146> 87 and m/z 146>92) exhibited similar distribution patterns, indicating that the correct distribution map for ACh was obtained. Conversely, the other fragment ions (84 and 72) exhibited a white matter-specific ion distribution. Obviously, the ISF sample provided the most sensitive ACh signal. (b) Expanded ACh distribution image of the hippocampus of an ISF-treated brain (left) and an optical image of the same brain section stained with H&E (right). For reference, corresponding brain atlas images are also shown (bottom). B. Quantitative imaging mass spectrometry (Q-IMS) analysis of the effects of therapeutic hypothermia on ACh. In the sham group, the [ACh]app values were high in the amygdala (Am) and the hippocampus, but relatively low in the cortex. Hypoxia-ischemia (H-I) caused substantial increases in [ACh]app in hippocampus and thalamus. Subsequently, a 3-hour reoxygenation under both normothermic and hypothermic conditions substantially decreased [ACh]app. Note that [ACh]app after normothermic treatment was lower than its pre-H-I values in many regions. (CL; contralateral, IL; ipsilateral.)
We further demonstrated that this method is sensitive enough to analyze dynamic changes in local ACh concentration between normal and diseased-conditioned brains in a model of neonatal cerebral ischemia.34) We showed that the local ACh concentration was significantly altered among control and neonatal ischemia-treated mice with normothermia or hypothermia, the latter of which showed brain region-specific reduction of ACh, suggesting a new mechanism of therapeutic hypothermia involving ACh metabolism.34)
After sampling of the animal organ with the optimized protocol(s) described above, the next key point to perform highly sensitive IMS is to enhance the target analyte ionization efficiency on the tissue surfaces, owing to the severe ion suppression effect. Since the tissue specimen is directly measured in IMS, it is important to consider that the samples are an extremely complex mixture of biomolecules. In typical ESI-MS measurements, the analyte molecules are extracted from such tissues and purified before ionization to achieve high sensitivity and selectivity. By contrast, in IMS, the sample cleanup procedure is limited to avoid molecular diffusion in the tissue sections. When such a crude sample is subjected to MALDI, numerous molecular species are competing for ionization, and, eventually, molecules that are abundant and/or easily ionized preferentially reach the detector. Consequently, these molecules suppress the ionization of other molecules (i.e., have an ion suppression effect), especially those present in trace amounts.50–52) Thus, when analyzing such trace signaling molecules, it is important to optimize the sample conditions so that the target molecule can be efficiently ionized from such a crude mixture.
To approach this problem, an on-tissue derivatization method, in which targeted metabolites are derivatized so that their chemical properties become altered to increase laser adsorption efficiency and/or to easily have an electric charge, has developed. This concept was first applied for the visualization of an exogenously administered drug, 3-methoxysalicylamine (3-MoSA), which is a scavenger of γ-ketoaldehydes53) (Fig. 4A). In this study, derivatization of this compound with 1,1-thiocarbonyldiimidazole resulted in an oxothiazolidine derivative, which could be detected with higher sensitivity by MALDI than the 3-MoSA itself. By expanding this concept, not only the exogenous drug but also the endogenous metabolites, including steroid hormones54) (Fig. 4B), minor amino acids,27) and mono-amine neurotransmitters along with their precursor/decomposed metabolites55,56) (Fig. 4C), were successfully visualized. Importantly, this derivatization method enables detection/visualization of trace metabolites existing at the nano molar level in vivo; for example, the monoamine transmitter dopamine (its brain concentration is less than a few nano molar57)) and the steroid hormone corticosterone (with an in vivo blood concentration less than micro molar58)).
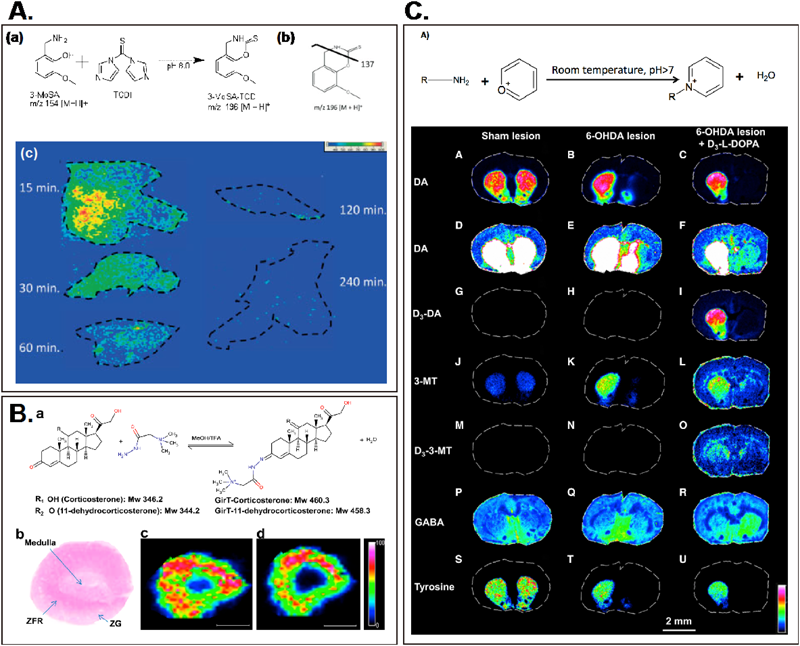
A. Reaction schema of 3-MoSA with TCDI (a), and fragmentation pattern of 3-MoSA–TCDI (b). (c) Time course MALDI-IMS for m/z 196>137 of liver tissue sections from dosed mice.53) B. MALDI-IMS results of corticosteroid derivatives prepared with Girard T reagent (GirT) in representative rat adrenal gland sections. Ion distribution maps showed GirT-CORT (m/z 460.31713) and GirT-11DHC (m/z 458.30139) in high abundance in the zona fasciculata/reticularis (site of glucocorticoids synthesis). (a) Derivatization of corticosterone and 11-dehydrocorticosterone with GirT. (b) Histological image of a cryosection of rat adrenal gland stained with hematoxylin and eosin (ZG=zona glomerulosa; ZFR=zona fasciculata reticularis).54) C. (a) Reaction of primary amines with pyrylium ion occurs at ambient temperature and pressure in 50% methanol solution at pH >7. Bottom: A–U) IMS were conducted on brain tissue sections from unilateral shamlesioned-, unilateral 6-OHDA-lesioned, and unilateral 6-OHDA-lesioned animals that were treated with subchronic L-DOPA for 4 weeks, with the final dose being given as deuterated (D3)-L-DOPA. The images show the distributions of DA (A–F) in sham-lesioned (A), 6-OHDA-lesioned (B), and 6-OHDA-lesioned L-DOPA-treated animals. Rescaling images (D–F) made it possible to determine the distribution of DA in the 6-OHDA-lesioned side of the brain and in structures with low DA concentrations, such as the cortex. Distribution of D3-DA (formed by the in vivo breakdown of D3-L-DOPA is imaged in shamlesioned (G), 6-OHDA-lesioned (H), and 6-OHDAlesioned L-DOPA-treated animals (I). Distribution of endogenous 3-MT is imaged in sham-lesioned (J), 6-OHDA-lesioned (K), and 6-OHDA-lesioned L-DOPA-treated animals (L), and distribution of D3-3-MT (derived from D3-L-DOPA) is imaged in sham-lesioned (M), 6-OHDA-lesioned (N), and 6-OHDA-lesioned L-DOPA-treated animals (O). Distribution of GABA (P–R) is imaged in shamlesioned (P), 6-OHDA-lesioned (Q), and 6-OHDAlesioned L-DOPA-treated animals (R), and tyrosine is imaged in sham-lesioned (S), 6-OHDA-lesioned (T), and 6-OHDA-lesioned L-DOPA-treated animals (U). There are clear differences in the concentrations of the studied amines in different regions of the brain and between treatments.55)
In Toue et al.,31) we describe a method for the tissue derivatization of amino acids and its application for research into cancer metabolism (Figs. 5A, B). Despite the abundance of major amino acids occurring at the micromolar order, to date, only a few endogenous amino acids have been detected by tissue MALDI-MS using conventional methods.9,23) This is presumably due to the low ionization efficiency of amino acids and matrix interference. By employing the TAHS derivatization method, which was initially developed for the ESI-MS technique, we demonstrated the successful detection and imaging of multiple amino acids in the tumor-bearing livers of immune deficient NOD/Shi-scid, IL-2R γ null (NOG) mice, a hepatic metastasis model for human colon cancer. After optimization of the derivatization conditions, reproducible and high-efficiency reaction rates were obtained for all protein-constituent amino acids, except for cysteine. By employing this TAHS derivatization method, Gly, Ala, Leu/Ile/HyPro, Asp, Gln, Glu, Phe, Tyr, and Trp were detected as TAHS derivatives in the liver tissue sections by MALDI-MS. Regarding detection sensitivity, TAHS derivatives of amino acids were detected by MALDI-MS at essentially the same order of magnitude as their measured concentrations in the liver sections. In addition, despite being present at low concentrations in the in vivo liver, the aromatic amino acids (Phe, Tyr, and Trp) were detected rather efficiently. This is presumably because these aromatic amino acids possess the ability to absorb ultraviolet light, which might cause them to exert “matrix” effects, suggesting that TAHS functions as a type of “reactive matrix.”56) In other words, the TAHS derivatization method provided an opportunity to analyze minor amino acids by improving their ionization efficiency, especially for aromatic amino acids.
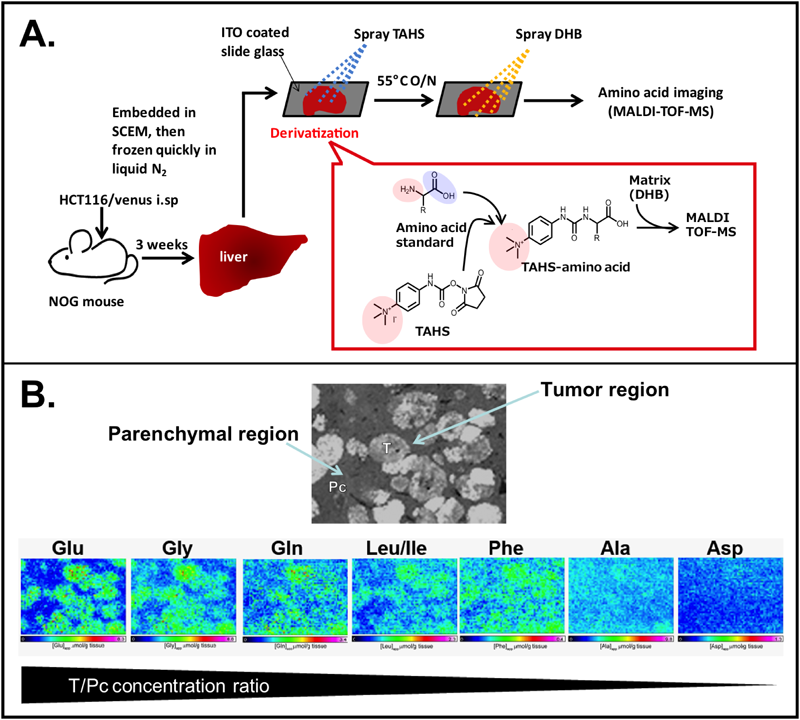
A. Schematic diagram of sample preparation for TAHS-derivatization. The human-derived colon cancer cell line HCT116/Venus was injected into the spleen of 16-week-old male NOG mice. Three weeks after the implantation, the liver tissues were removed to be snap-frozen. The frozen tissue sections were prepared and derivatized with TAHS to serve as samples for MALDI-IMS. B. Representative MS images of Glu-TAHS, Gln-TAHS, Gly-TAHS, Leu/Ile/HyPro-TAHS, Phe-TAHS, Asp-TAHS, and Ala-TAHS in the tumor-bearing livers are arranged in order of those tumor accumulation levels. Optical images show the local- ization of metastatic tumors derived from Venus-expressing HCT116 cells.
Another important advantage of on-tissue TAHS derivatization is that it enables the simultaneous imaging of multiple amino acids under the same experimental condition. In the conventional methods for MALDI-IMS of amino acids, it is necessary to select an appropriate matrix and a positive/negative ion detection mode based on the chemical properties of each amino acid.9,23) In this regard, TAHS derivatization simplifies these experimental procedures by providing cationic charges to all amino acids. Consequently, these cationically charged amino acid derivatives can be simultaneously detected by use of 2,5-dihydroxybenzoic acid in positive-ion mode. The imaging results presented in Fig. 5B revealed a distinct amino acid metabolism pattern in tumors and the host parenchymal regions. Application of our method to the investigation of human cancer xenografts deserves further study to elucidate the mechanisms underlying the interplay of amino acid metabolism between the cancer and host tissue in vivo.
In order to understand biochemical basis of disease progression and to develop new therapeutic strategies, it is important to characterize the spatiotemporal changes of bioactive small molecules in vivo. This technical task was accomplished by developing a highly sensitive IMS technology. Although MALDI-IMS has powerful capabilities for visualizing many metabolites in discrete areas, the sensitivity and selectivity of IMS is still insufficient. Therefore, such characterization requires a technical breakthrough, including a highly sensitive IMS technique that is capable of detecting low levels of metabolites in the heterogeneous regions of abnormal tissues in diseased organs. As reviewed here, our group and others have tackled this challenge by (i) optimization of the sample preparation protocol, and (ii) the use of on-tissue derivatization in combination with MALDI-IMS. This work highlights that these approaches enable imaging of signal transmitters at sub-nano molar-level in tissues. Further investigations are required, especially those that focus on developing more derivatization reagents that are optimized for MALDI-IMS, such as a reactive matrix. On the other hand, we believe that these promising approaches will help to answer important unresolved questions in disease progression, such as determining the specific regions of organs or cells in vivo from which abnormal inflammatory mediators are produced. Resolving these issues would provide the key to understanding the biological basis of chronic inflammatory diseases, discovering new diagnostic markers, and developing new therapies.