2017 Volume 93 Issue 5 Pages 339-359
2017 Volume 93 Issue 5 Pages 339-359
Mitochondrial DNA (mtDNA) is packaged by association with specific proteins in compact DNA-protein complexes named mitochondrial nucleoids (mt-nucleoids). The budding yeast Saccharomyces cerevisiae is able to grow either aerobically or anaerobically. Due to this characteristic, S. cerevisiae has been extensively used as a model organism to study genetics, morphology and biochemistry of mitochondria for a long time. Mitochondria of S. cerevisiae frequently fuse and divide, and perform dynamic morphological changes depending on the culture conditions and the stage of life cycle of the yeast cells. The mt-nucleoids also dynamically change their morphology, accompanying morphological changes of mitochondria. The mt-nucleoids have been isolated morphologically intact and functional analyses of mt-nucleoid proteins have been extensively performed. These studies have revealed that the functions of mt-nucleoid proteins are essential for maintenance of mtDNA. The aims of this review are to summarize the history on the research of yeast mt-nucleoids as well as recent findings on the organization of the mt-nucleoids and mitochondrial dynamics.
Mitochondria are indispensable organelles that serve as power plants of eukaryotic cells because the majority of ATP required for activity of cells is produced by the TCA cycle and oxidative phosphorylation in mitochondria. In addition to ATP synthesis by respiration, mitochondria gained a variety of functions, like calcium ion homeostasis, heme synthesis, lipid synthesis and apoptosis, during the evolution of eukaryotic cells.
Mitochondria in aerobically respiring cells maintain their own genome, mitochondrial DNA (mtDNA), which is reminiscent of evolution from ancient intracellular symbionts that belong to α-proteobacteria. As mitochondria are the major source of reactive oxygen (ROS), which is the byproduct of respiration and toxic for the cell, mtDNA is thought to be more susceptible to oxidative damage than nuclear DNA. Nuclear DNA is tightly folded and protected in ordered nucleosome structures by association with histones. However, mitochondria do not contain histones, and mtDNA has been for long time considered to be free from protective proteins and naked. Kuroiwa et al.1) demonstrated that mtDNA in microplasmodia of the slime mold, Physarum polycepharum, is localized to electron dense bodies, called mitochondrial nuclei or nucleoids at the central region of mitochondria. They succeeded to isolate the mitochondrial nucleoids (mt-nucleoids) keeping their intact structure, and showed that mtDNA is three-dimensionally organized with specific proteins.1) These studies established the concept of mt-nuclei (mt-nucleoids) that mitochondrial division accompanies division of mt-nucleoids during the mitochondrial division cycle. Thus, the mt-nucleoid structures are essential for correct replication, maintenance and inheritance of mitochondrial genome.
The baker’s yeast, Saccharomyces cerevisiae, has long been a model organism for research on mitochondrial genetics. Therefore, we consider that in order to confirm the generality of the structure of mt-nucleoids, it is important to reveal the structure and function of the mt-nucleoids in yeast cells. There are excellent reviews describing the organization of yeast mt-nucleoids.2),3) In this review, first, we will briefly summarize the history of yeast mitochondrial genetics and morphology. Next, we will describe the research on morphology and molecular organization of the mt-nucleoids including our research. Finally, we will describe the recent findings on distribution of the mt-nucleoids accompanying mitochondrial dynamics.
In the 1940’s, before the discovery of mtDNA, Boris Ephrussi, a French Geneticist, chose baker’s yeast, S. cerevisiae, as an organism for his genetic research. He found that petite colonies occurred on agar plates after treatment of yeast cells with mutagens like acriflavine.4) Petite colonies grew on fermentative carbon sources, but did not on non-fermentative carbon sources. This indicated that the petite colonies have the respiratory-deficient characteristics. The occurrence of petite colonies reached to nearly 100% after prolonged treatment with a high concentration of ethidium bromide, and unexpectedly, segregation of the petite phenotype did not follow Mendelian inheritance. He speculated that an unknown cytoplasmic factor named rho determines the respiration activity. Thus, wild-type cells are called rho+ and respiratory-deficient cells are called rho− or rho0.
The mtDNA of the yeast S. cerevisiae has a circular genetic map of 75–85 kbp and encodes 7 proteins for oxidative phosphorylation, one mitochondrial ribosomal protein, 2 rRNAs, 24 tRNAs, the 9S RNA component of RNase P and several intron-related open reading frames (ORFs).5) Treatment of rho+ cells with ethidium bromide resulted in large deletion of mtDNA, and a part of mtDNA fragments can survive and replicate as circular DNA in rho− cells. Cells that completely lack mtDNA were called rho0 cells. Due to the essential roles of mitochondria, except for respiration, mitochondrial structures are persistently maintained even in rho− and rho0 cells. The occurrence of rho− and rho0 cells is a characteristic feature of S. cerevisiae.
Mitochondrial genetics of yeast cells has developed using crosses among rho+, rho− and rho0 strains. Crosses with rho+ and rho− cells results in heteroplasmy, which means a mixture of two different types of mtDNA in a zygote. Following multiple budding up to 20 generations, each cell contains only a single species of mtDNA from either parent, which is called homoplasmy. The ratio of descendants that maintain rho+ mtDNA or rho− mtDNA varies depending on the character of rho− mtDNA. In a cross of rho+ and rho− cells, specific rho− mtDNA is predominantly transmitted to descendants. This segregation is called hypersuppressiveness.6) MtDNA that shows hypersuppressiveness contains multiple repeats of the replication origin (ori) sequence. It has been explained that selective replication and transmission of the ori sequence will lead to hypersuppressiveness, but the correct mechanisms are not fully understood. Genetic studies with antibiotic resistant mitochondrial markers demonstrated a high ratio of recombination events between yeast mtDNAs.7)
A yeast cell contains 50–100 copies or more of mtDNA depending on strains. However, there is a contradiction called “ploidy paradox” between the large physical number of mtDNA molecules per cell and the small number (1–5) of heritable units, which were estimated from mitochondrial genetics.8) In order to understand the results obtained by mitochondrial genetics, it was essential to visualize mtDNA in the cells.
The yeast, S. cerevisiae, has been an attractive organism for the study of mitochondrial biogenesis because S. cerevisiae can grow either aerobically or anaerobically. At the start of the research, electron microscopic observations suggested that the mitochondria change their morphology from large structures to small ones according to the progress of vegetative growth.9) Hoffmann and Avers10) reported by three-dimensional reconstruction of serial ultrathin sections that mitochondria in a yeast cell are continuous to form a single organelle. Stevens11),12) has also performed detailed electron microscopic observation of yeast mitochondria and reported that the number, shape and volume of mitochondria varies in close relation to the life cycle and physiological state of the cells. She revealed that the log-phase cells contain few tubular mitochondria, but stationary-phase cells contain many spherical or fragmented ones. This means the morphology of mitochondria is not static but dynamically changing by response to environmental conditions. Throughout the electron microscopic observations, mtDNA was observed as tiny filamentous structures in electron-transparent areas in ultrathin sections of mitochondria. It has not been well understood how mtDNA is three-dimensionally organized in yeast mitochondria.
Detailed quantitative analyses of tubular mitochondrial network were done using beam-scanning multifocal multiphoton 4Pi microscopy in live cells.13) These studies showed that mitochondria in glycerol-grown cells exhibit a strongly branched tubular reticulum as opposed to glucose-grown cells. The average tubular diameter was 339 nm in glucose-grown cells, whereas it increased to 360 nm in glycerol-grown cells. This change resulted in a 2.8-fold increase of surface area and 3.0-fold increase in volume of mitochondria. These results coincided with the detailed observations of mitochondrial structure by Stevens.12)
A DNA-binding fluorochrome, 4′,6-diamidino-2-phenylindole (DAPI) was first used for visualization of yeast mitochondrial DNA.14) The DAPI-staining technique was successfully used to reveal that mtDNA of slime mold, Physarum polycepharum, is three-dimensionally organized with specific proteins to form electron-dense rod-like mt-nucleoids.1)
Using the DAPI-staining technique, we demonstrated that yeast mt-nucleoids appeared as strings-of-beads in tubular mitochondria in log-phase cells, but as many fluorescent dots in stationary-phase cells.15) The mt-nucleoids seemed to fuse to form strings-of-beads during meiosis and sporulation in a network of tubular mitochondria. In those observations, the rigid cell wall prevented clear observation of the mt-nucleoids. Subsequently, in order to observe the morphology of the mt-nucleoids more clearly, we developed a synchronous sporulation culture of spheroplasts.16) This method included the fractionation of large cells that perform synchronous sporulation and spheroplast formation by Zymolyase to digest the cell wall.16),17) Observation of spheroplasts markedly improved fluorescence images of the mt-nucleoids by DAPI staining (Fig. 1). Cells that reached the stationary phase contained approximately 60 discrete mt-nucleoids stained with DAPI (Fig. 1A). When spheroplasts were inoculated in sporulation medium, all the mt-nucleoids lined up as strings-of-beads and coalesced to a network of mt-nucleoids that enclosed the cell periphery (Fig. 1B). The network of mt-nucleoids changed their localization from the cell periphery to the vicinity of dividing nuclei at the stage of meiotic division I and II (Fig. 1C). Finally, the network of mt-nucleoids formed a ring structure that surrounded each spore nuclei (Fig. 1D). These unique distributions of mt-nucleoids reflect the morphological changes from fragmented mitochondria to a tubular network, as revealed by 3,3′-dihexyloxacarbocyanine iodide [DiOC6(3)] staining of mitochondria. We also demonstrated that mitochondria dynamically fuse to form a network during germination of spores, budding, and zygote formation.16),18) Our findings indicated that unknown mechanisms function for effective inheritance of mitochondria into spores. However, before 1990, nothing was known about how mitochondrial morphology was regulated.
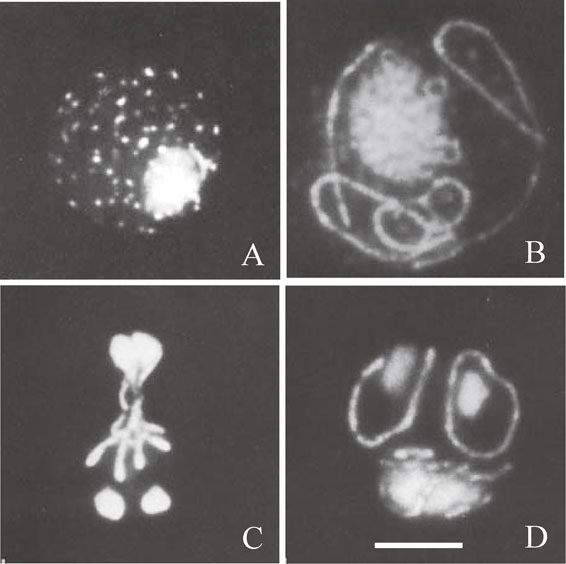
Fluorescence photomicrographs showing cell nuclei and mt-nucleoids in spheroplasts during meiosis and sporulation after DAPI staining. (A) In stationary-phase cells, mt-nucleoids appeared as fluorescent dots. (B) In cells at meiotic prophase, mt-nucleoids appeared as strings or strings-of-beads. (C) In cells after the second meiotic division, the strings of mt-nucleoids produced X-shaped morphology in the central region of four nuclei. (D) In cells just before spore wall formation, a ring of mt-nucleoids encloses each daughter nucleus. Bar represents 5 µm. A–D is from Ref. 16.
Development of synchronous sporulation culture of spheroplasts gave us an unexpected chance to observe meiotic chromosomes. At that time, the number of linkage groups had been predicted to be 16 or 17 in S. cerevisiae from genetic crosses. However, direct visualization of yeast chromosomes was difficult under optic microscopy because the genome size of S. cerevisiae is only 12 Mbp and its value is about one 250th of the human genome. Therefore, different chromosome numbers have been reported. Fortunately, we have successfully observed the process of chromosome condensation during meiotic prophase with DAPI staining. (Ref. 16, Fig. 1) We confirmed for the first time the karyotype of 16 bivalent chromosomes during meiotic prophase.19),20) DNA content of each chromosome was well coincided to the value estimated from genetic map.
Electron microscopy demonstrated that mitochondria of Physarum polycephrum have distinct electron dense mt-nucleoids.1) On the other hand, yeast mtDNA has been shown to be filamentous structures located in the transparent area in the matrix of mitochondria, as observed by electron microscopy.12) Therefore, we asked a fundamental question of whether yeast mtDNA is also three-dimensionally organized into nucleoid structures to be associated with proteins. In order to disrupt cells in mild conditions, spheroplasts were prepared by digestion of the cell wall with Zymolyase. The protocol of mt-nucleoid isolation includes disruption of spheroplasts, the isolation of mitochondria with differential centrifugations and the isolation of mt-nucleoids from mitochondria with sucrose density centrifugations. We demonstrated that the mt-nucleoids could be isolated from spheroplasts of the stationary phase cells, maintaining their size that appeared in vivo.21) Measurement of fluorescence intensity of DAPI-stained mt-nucleoids showed that the average number of mtDNA molecules in an mt-nucleoid was estimated as 3.9 in spheroplasts and 3.1 in isolated mt-nucleoids. Electron microscopic observation indicated that the isolated mt-nucleoids have chromatin-like structures associated with proteins. These results demonstrated that the yeast mtDNA is not naked, but packaged into nucleoid structures associated with proteins, although the electron dense areas of the mt-nucleoids were not observed in yeast mitochondria by electron microscopy. We detected a number of proteins in the mt-nucleoid fractions. Among them, the presence of a unique histone-like protein of 20 kDa with basic pI was evident on acid-urea two-dimensional electrophoresis.21)
At that time, a histone-like DNA-binding protein named HM, which induced negative supercoil to circular DNA in the presence of topoisomerase I, had been isolated from yeast mitochondria.22) The molecular weight and basic pI of the 20-kDa protein that we detected in the isolated mt-nucleoids well coincided with those of HM. The gene coding for HM was cloned and designated as ABF2.23) Abf2 is encoded by a nuclear gene and contains two DNA-binding domains, HMG boxes that have sequence similarity to the non-histone protein HMG-1. Abf2 is an abundant protein present at a ratio of one molecules of Abf2 for every 15 bp of mtDNA.23)
As shown in Fig. 2A, Abf2 is a major mt-nucleoid protein, but a number of proteins are also associated with the mt-nucleoids.24) The isolated mt-nucleoids were disassembled to mtDNA and proteins by the addition of 2 M NaCl. The disassembled mt-nucleoids were reassembled once again into compact structures by decreasing the NaCl concentration by dialysis.24) DNA-cellulose chromatography of the mt-nucleoid proteins demonstrated that Abf2 were eluted from the column at 0.6 M NaCl, but the other five species of the mt-nucleoid proteins were eluted from the column at 0.2 M NaCl. This indicated that Abf2 is an mt-nucleoid protein with the highest affinity to double-stranded DNA, but several other DNA-binding proteins bind to mtDNA with lower affinity. We showed that Abf2 folded DNA into chromatin-like structures, but the packaging of mtDNA is not very tight. Unexpectedly, mtDNA-binding proteins with low affinity to DNA packaged mtDNA more tightly, indicating that mt-nucleoid proteins other than Abf2 have also served to package mtDNA into chromatin-like structures.24)
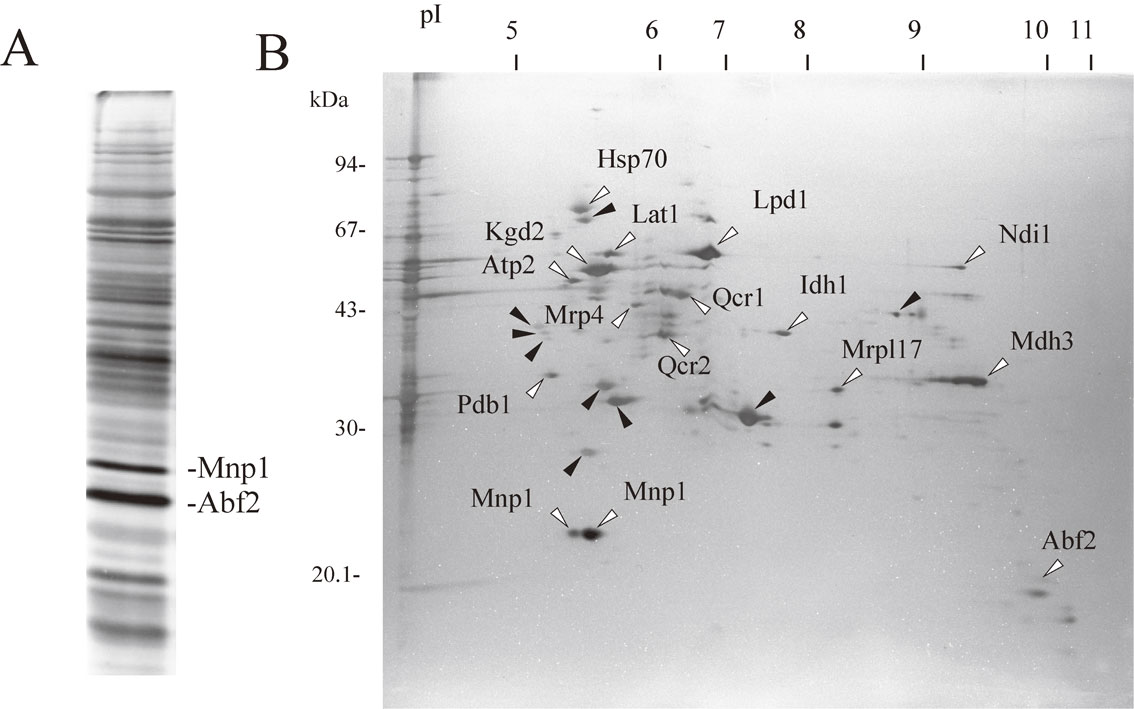
Separation of the mt-nucleoid proteins. (A) The mt-nucleoids were isolated from S. cerevisiae strain G2-2 grown to stationary phase in modified Burkholder’s medium. The mt-nucleoid proteins were separated by SDS-PAGRE and silver-stained. Two proteins of Abf2 and Mnp1 are indicated. (B) The mt-nucleoid proteins were separated by two-dimensional gel electrophoresis and silver-stained. Proteins were transferred to PVDF membrane. The membrane was stained with amido black. Each spot of the protein was excised, and the N-terminal amino acid sequence of the protein was determined with an Applied Biosystems Procise sequencer or Shimadzu protein sequencer PPSQ-21A. Proteins indicated by white arrowheads are the identified proteins, and proteins indicated by black arrowheads are the unidentified proteins. Figure 2A and 2B are from Ref. 24 and Ref. 39, respectively.
Abf2 introduces negative supercoils to circular DNA in the presence of topoisomerase I. This means that a major role of Abf2 is the packaging of mtDNA into the nucleoid structure. This is similar to the role of histones in packaging nuclear DNA into nucleosomes. Deletion of the ABF2 gene (Δabf2) leads to loss of mtDNA in cells grown in the presence of glucose as a carbon source, but not in cells grown in the presence of glycerol as a carbon source.23) Instability of mtDNA due to loss of Abf2 can be complemented by expression of nuclear HMG protein NHP6A targeted to mitochondria.25) Interestingly, the E. coli DNA-binding protein HU, which is targeted to mitochondria, can also complement the function of Abf2, although E. coli HU is not a HMG box protein.26) A mitochondrial transcription factor named h-mtTFA/TFAM is a homolog of yeast Abf2 (Fig. 3). TFAM also has two HMG domains and functionally replaces Abf2.27)
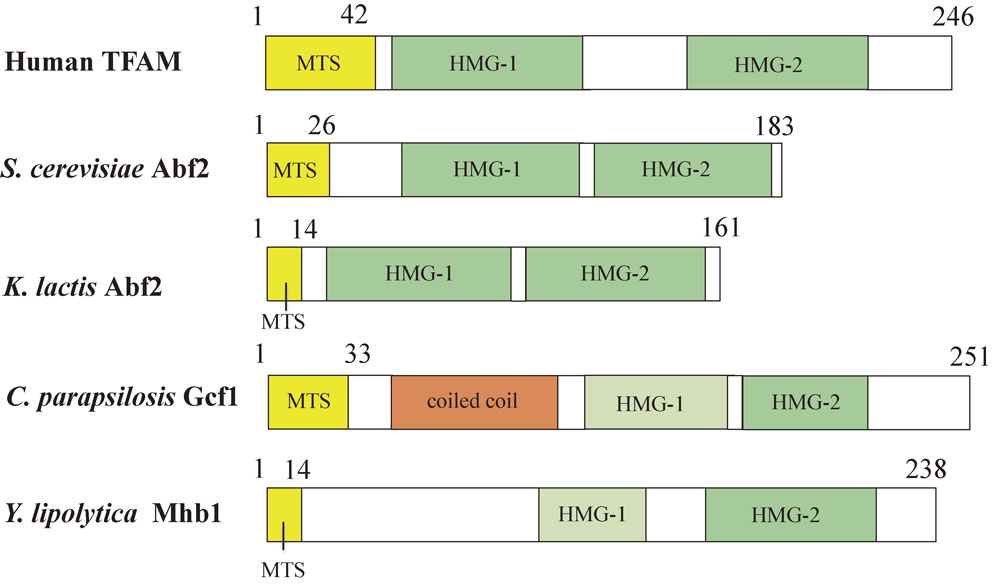
Comparison of mitochondrial HMG box proteins. The mitochondrial targeting sequence (MTS) is shown in yellow. HMG box domains detected by SMART are shown in green. HMG box domains that are not detected by SMART, but detected by the Phyre server are shown in pale green. HMG box1 of C. parapsilisis Gcf1 and Y. lipolytica Mhb1 is weakly conserved and considered to diverge from the typical amino acid sequence of HMG boxes. The coiled coil domain is shown in orange. Human TFAM, S. cerevisiae Abf2, K. lactis Abf2, C. parapsilosis Gcf1, Y. lipolytica Mhb1 are from Ref. 27, Ref. 23, Ref. 69 and 71, Ref. 76 and Ref. 78, respectively.
The functions of Abf2 have been investigated in detail in relation to mtDNA segregation, recombination and copy number control. Unlike mammalian TFAM that has a role in transcription, Abf2 does not seem to directly interact with the transcription machinery. The mtDNA content of rho+ cells is reduced by 70% in the Δabf2 cells grown in YPG medium containing glycerol as a carbon source.28) In addition, increasing the gene dosage of ABF2 by two to three fold resulted in a 50–150% increase in the amount of mtDNA per cell.28) This indicates that Abf2 is involved in regulation of the mtDNA copy number. In contrast, overexpression of the ABF2 gene resulted in instability of mtDNA. In homozygous Δabf2 crosses, mtDNA recombination is suppressed relative to homozygous ABF2 crosses.28) Indeed, it has been demonstrated by two-dimensional agarose gel electrophoresis that Abf2 promotes or stabilizes Holliday recombination junction intermediates in rho+ mtDNA in vivo.29) These results show that Abf2 functions in the recombination of mtDNA as well as in packaging of mtDNA.
It has been reported that the mt-nucleoids in Δabf2 cells are not compactly packaged but diffused, and have the protein composition that is different from that of wild-type cells.30) On the other hand, our observation showed that the mt-nucleoids appeared as discrete dots even in Δabf2 cells that are similar to mt-nucleoids in wild-type cells.31) We isolated the mt-nucleoids from Δabf2 cells grown in glycerol medium. Comparison of the mt-nucleoid proteins by SDS-PAGE and two-dimensional gel electrophoresis demonstrated that the protein composition of Δabf2 mt-nucleoids is not largely altered, except for lack of Abf2.31) Thus, it is still controversial as to whether the lack of Abf2 significantly changes the overall protein composition of mt-nucleoids. The number of mt-nucleoids is significantly decreased in Δabf2 cells grown in medium containing glycerol as a carbon source.31) Additionally, we noticed that the number and size of mt-nucleoids transmitted to the small bud is more markedly decreased in Δabf2 cells than in wild-type cells, although mitochondria is normally transmitted to the bud even in Δabf2 cells.31) As described later, mtDNA replication is considered to be mediated by the recombination process. Therefore, our results suggest that Abf2 is involved in transmission of mt-nucleoids by recombination-mediated mtDNA replication.
Abf2 nonspecifically binds to DNA, but exhibits a preference for GC-rich sequences.32) The process of DNA packaging by Abf2 was extensively analyzed in vitro with high-resolution atomic electron microscopy.33),34) Abf2 binding was reported to induce drastic bends by ∼78° in the DNA backbone for both linear and circular DNA, and at a high concentration of Abf2, DNA collapses into a tight nucleoprotein complex similar to mt-nucleoids. Abf2 was demonstrated to compact DNA through a simple mechanism that involves bending of the DNA backbone. However, Abf2 is not strongly bound to DNA compared with the histones.33),34)
Recently, Chakraborty et al.35) solved the crystal structures of Abf2 in complex with mtDNA-derived fragments. They revealed that each HMG box of Abf2 induces 90° bend in the contacted DNA, causing an overall U-turn. They also demonstrated that a characteristic N-terminal flag and helix of Abf2 is crucial for mtDNA maintenance.
A number of proteins were detected in the isolated mt-nucleoid fractions in addition to Abf2 (Fig. 2A). Attempts to identify the mt-nucleoid proteins have been performed in several laboratories. These approaches included production of monoclonal antibodies against the mt-nucleoid proteins,36),37) analysis of the mt-nucleoid proteins by two-dimensional gel electrophoresis38),39) and formaldehyde crosslinking of mt-nucleoid proteins to mtDNA.40) We separated the mt-nucleoid proteins by two-dimensional gel electrophoresis and determined the amino acid sequence of each protein (Fig. 2B). A list of candidates for the mt-nucleoid proteins determined by us and other laboratories is shown in Table 1. We describe characteristics of several important proteins below, and the predicted image of the mt-nucleoid structure is illustrated in Fig. 6A.
| Protein | Primary function | Reference |
|---|---|---|
| Abf2 | HMG box protein, DNA Packaging | 39, 40, 44 |
| Rim1 | Single-stranded DNA-binding | 40, 44 |
| Mgm101 | mtDNA maintenance and repair | 40, 44 |
| mtHsp70 (Ssc1) |
Mitochondrial chaperonin | 39, 44 |
| mtHsp60 | Mitochondrial chaperonin | 40, 42 |
| Ilv5 | Acetohydroxy acid reductoisomerase | 38, 44 |
| Kgd1 | α-ketoglutarate dehydrogenase subunits | 37, 39, 44 |
| Kgd2 | α-ketoglutarate dehydrogenase subunits | 37, 39, 40, 44 |
| Lpd1 | α-ketoglutarate dehydrogenase subunits | 37, 37, 40 |
| Lat1 | Pyruvate dehydrogenase subunits | 39 |
| Pda1 | Pyruvate dehydrogenase subunits | 44 |
| Pdb1 | Pyruvate dehydrogenase subunits | 39, 44 |
| Ald4 | Mitochondrial Aldehyde dehydrogenase | 40, 44 |
| Aco1 | Aconitase | 40, 44 |
| Atp1 | F1-ATPase subunit | 40, 44 |
| Atp2 | F1-ATPase subunit | 39 |
| Ndi1 | NADH-ubiquinone-6 oxidoreductase | 39 |
| Qcr2 | Ubiquinol-cytochorome c reductase | 39 |
| Qcr1 | Ubiquinol-cytochorome c reductase | 39 |
| Mrp4 | Mitochondrial ribosomal protein | 39 |
| Mrpl17 | Mitochondrial ribosomal protein | 39 |
| Mnp1/bL12 | Mitochondrial ribosomal protein | 39, 54 |
| Mdh3 | Malate dehydrogenase | 39 |
| Rpo41 | RNA polymerase | 44 |
| Sls1 | Mitochondrial translation | 44 |
| mtHsp40 (Mdj1) |
Mitochondrial chaperonin | 117 |
| mtHsp10 | Mitochondrial chaperonin | 44 |
| Ilv6 | Acetolactate synthase regulatory subunit |
44 |
| Idh1 | NAD+-dependent isocitrate dehydrogenase |
44 |
| Idp1 | NADP+-dependent isocitrate dehydrogenase |
44 |
| Cha1 | L-serine/L-threonine deaminase | 44 |
| Lsc1 | Succinate-CoA ligase α subunit | 44 |
| Arg5,6 | Arginine biosynthesis | 118 |
| Mip1 | DNApolymerase gamma | 49 |
| Yhm2 | DNA-binding protein | 64 |
Zeleneya-Troitskaya et al.41) identified a multicopy suppressor of the mtDNA instability phenotype of Δabf2 cells. Surprisingly, the suppressor, IlV5 encoded acetohydroxy acid reductoisomerase, which catalyzes a step in the biosynthesis of the branched-chain amino acid, isoleucine and valine. Deletion of the ILV5 gene (ΔIlv5) leads to high frequency of petite mutation, and the stability of mtDNA is severely impaired in Δabf2 Δilv5 double mutants. The ilv5 mutants that lack either enzymatic activity or the DNA-binding activity were isolated.42) Although rho+ mtDNA was stably maintained in cells with catalytically inactive Ilv5, the ilv5 mutant that lost DNA-binding activity produced petite mutation. This indicated that DNA-binding activity of Ilv5 is required for maintenance of mtDNA. Ilv5 directly binds double-stranded DNA in vitro, and was co-fractionated with mtDNA and Abf2 when the mt-nucleoids were loaded on sucrose density gradient centrifugations.43) Interestingly, the Ilv5/mtDNA ratio increased 10-fold in the mt-nucleoids isolated from Δabf2 cells in comparison to the Ilv5/mtDNA ratio in the wild-type mt-nucleoids.43)
Aconitase (Aco1p).Aconitase is a matrix enzyme that catalyzes the conversion of citrate to isocitrate in the TCA cycle. Chen et al.44) revealed that deletion of the ACO1 gene that codes for aconitase (Aco1) leads to the loss of mtDNA. The overexpression of wild-type Aco1 and the mutant Aco1 that lacks enzymatic activity can suppress the mtDNA instability seen in Δabf2 cells. Thus, Aco1 is required for mtDNA stability, independent of its catalytic activity. Aco1 has the ability to bind DNA in vitro, and its DNA-binding activity is required for mtDNA maintenance in vivo.45) Aco1 is co-fractionated with mtDNA by sucrose density centrifugations of mt-nucleoids.32) This suggests that Aco1 can replace the mtDNA packaging function of Abf2. Thus, aconitase is a multi-functional enzyme that has two different regions, a DNA-binding site and a catalytic site.
On the other hand, an alternative mechanism has been recently proposed to account for mtDNA loss in aconitase deficient (Δaco1) cells.46) The deletion of the ACO1 gene activates the retrograde response pathway (RTG pathway) in response to defects in mitochondrial respiration. The RTG pathway activates the expression of genes (CIT1, CIT2, CIT3) that code for citrate synthase, which induced accumulation of citrate in mitochondria. The increase in the citrate level in turn increased the iron concentration, which converts superoxide radicals to hydroxyl radicals that are more damaging to mtDNA than superoxide radicals. Consequently, the deletion of ACO1 induced the loss of mtDNA. This mechanism is different from the mechanism previously proposed by Chen et al.,44) in which aconitase has a direct role in physical protection of mtDNA. The two different mechanisms may account for the role of aconitase in mtDNA stability.
MGM101.Chen et al.47) selected a temperature sensitive mutant named mgm101 that could not maintain mtDNA at restriction temperature of 37 °C. Subsequently, it was found that Mgm101 is exclusively associated with the mt-nucleoids and binds to DNA. Mgm101 is required for the repair of oxidative mtDNA damage.48) Interestingly, Mgm101 is associated with a subpopulation of mt-nucleoids, in which mtDNA replication occurs.49) Recent research demonstrated that Mgm101 preferentially binds to single-stranded DNA and catalyzes the annealing of single-stranded DNA pre-complexed with the mitochondrial single-stranded DNA-binding protein, Rim1. Mgm101 is related to the Rad52-type recombination proteins.50)
The Mgm101 interacts with MuSα and the DNA helicase Mph1, and is required for nuclear DNA interstrand cross-link (ICL) repair in the absence of Pso2 exonuclease in S. cerevisiae.51) Cellular fractionation of the nuclear and mitochondrial compartments confirmed that Mgm101-FLAG was present in both fractions.51) It is suggested that Mgm101 functionally overlap with Rad52 and is required for telomere elongation during chromosome replication in rad52 defective cells.52) There are interspecific differences in biochemical properties of Mgm101. The CpMgm101 in the yeast Candida parapsilosis maintaining a linear mtDNA is associated with the entire mt-nucleoid population.53) DNA-binding properties of CpMgm101 suggest that it can play a number of possible roles in the replication of the mitochondrial genome and maintenance of its telomeres.53)
Mnp1p.We detected two spots of 22-kDa proteins, which were highly concentrated in the mt-nucleoids fraction, with acidic pI by two dimensional gel electrophoresis of the isolated mt-nucleoids (Ref. 54, Fig. 2AB). The N-terminal amino acid sequence of both proteins coincided with an unidentified protein (systematic name YGL068W) that is predicted to be a mitochondrial ribosomal protein homologous to the ribosomal proteins L7/L12 of E. coli. We named this protein Mnp1. Two isoforms of Mnp1 with different pIs suggest some chemical modifications. We confirmed that Mnp1 is localized to the mt-nucleoids by immunofluorescence microscopy and DAPI-staining. Lack of Mnp1 resulted in instability of mtDNA that led to petite mutation. Therefore, Mnp1 is required for maintenance of mtDNA. The yeast mitochondrial ribosomal protein L7/L12 is supposed to be localized to the surface exposed region of the mitochondrial large ribosomal subunit.55) The association of mt-nucleoids and the mitochondrial ribosome may be possible because translation is coupled with transcription in mitochondria. Alternatively, it is also possible that Mnp1 has a role for stabilizing mtDNA, independent of the mitochondrial ribosome. Interestingly, Mnp1 has not been yet identified from the isolated mitochondrial ribosome by mass spectrometric analysis.56)–58) However, this protein can be easily detected as a relatively major protein of the mt-nucleoids (Fig. 2AB). Mnp1 has not been detected by formaldehyde crosslinking of proteins to mtDNA.40),44) The detailed function of Mnp1 remains to be determined. It has been reported that a human mitochondrial ribosomal protein MRPL21, which is a homolog of E. coli L7/L12, binds to mitochondrial RNA polymerase independent of the mitochondrial ribosome.59)
Subunits of α-ketoglutarate dehydrogenase.In order to identify the components of mt-nucleoids, we tried to raise monoclonal antibodies against the isolated mt-nucleoids and produced one monoclonal antibody named YMN-1.36) The YMN-1 antibody binds to the mt-nucleoids in cells or the isolated mt-nucleoids as revealed by immunofluorescence microscopy. The antigen protein of the YMN-1 monoclonal antibody was dihydrolipoyl transsuccinylase, Kgd2, a subunit of α-ketoglutarate dehydrogenase (KGDC) that catalyzes the oxidative decarboxylation of α-ketoglutarate to succinyl-CoA in the TCA cycle.37) We obtained two other components of KGDC, namely, α-ketoglutarate dehydrogenase (Kgd1) and dihydrolipoyl dehydrogenase (Lpd1) in DNA-binding protein fractions after DNA-cellulose chromatography of disassembled mt-nucleoid proteins.37) Formaldehyde-crosslinking of mt-nucleoids also detected Kgd2 and Lpd1.40) Genetic experiments demonstrated that deletion of the KGD2 gene increased the petite-inducing phenotype of the Δabf2 mutant.40) This suggests that Kgd2 participates in the stability of yeast mtDNA.
HSP60.Hsp60 has been identified as the mt-nucleoid protein, which binds preferentially to the template strand of ori5 DNA, a putative replication origin of mtDNA.40) Genetic experiments indicated that temperature-sensitive hsp60 mutations cause petite mutation. This shows that Hsp60 also participates in the stability of mtDNA. It has been suggested that mtDNA instability in Δhsp60 cells is caused by a defect in mtDNA transmission to daughter cells.60) Hsp60 is recruited to mt-nucleoids in glucose-grown cells during glucose repression.32)
Although a number of proteins have been discovered as mt-nucleoid proteins as shown in Table 1, it should be noted that careful investigation is needed to for identification of mt-nucleoid proteins. The proteins, whose overexpression suppresses the instability of mtDNA in Δabf2 cells, are not necessarily the mt-nucleoid proteins. The SHM1 gene, which was renamed to GGC1/YHM1, has been identified as a multicopy suppressor of the temperature-sensitive defect associated with the Δabf2 mutation, although Shm1 did not exhibit DNA-binding activity.61) On the other hand, it has been reported that the deletion of the SHM1/GGC1/YHM1 gene resulted in iron accumulation in mitochondria.62) According to the proposed mechanism, iron accumulation in Δyhm1 cells produces toxic free-radicals that will lead to DNA damage.62) Overexpression of the SHM1/GGC1/YHM1 gene in Δabf2 cells may reduce the iron concentration in mitochondria, which will in turn decrease the concentration of free-radicals and result in stabilization of mtDNA. It has been also reported that the SHM1/GGC1/YHM1 gene codes for a GTP/GDP transporter in the mitochondrial inner membrane.63) Another multicopy suppressor (YHM2) of mtDNA instability associated with the Δabf2 mutation has been identified. Yhm2 is localized in the mitochondrial inner membrane and associated with the isolated mt-nucleoids in vitro.64) Later, biochemical characterization of purified Yhm2 revealed that Yhm2 is a mitochondrial inner membrane protein that serves as transporter for citrate and oxoglutarate.65)
A number of mitochondrial proteins have also been classified as the mt-nucleoid proteins of mammals according to mass-spectrometry based analysis.66) Hansen et al.66) noted that some proteins involved in replication, transcription, translation and repair may attach transiently to mt-nucleoids, but other proteins may stably bind to the mt-nucleoid proteins as structural components. Additionally, the composition of mt-nucleoids may largely depend to the methods to isolate the mt-nucleoids. This situation is also true for the identification of yeast mt-nucleoid proteins.
Using SDS-DNA polyacrylamide gel electrophoresis (SDS-DNA PAGE) that was developed for detection of nuclease and nuclear histones, we were able to specifically detect Abf2 among the mt-nucleoid proteins on gels. This method includes separation of mt-nucleoid proteins on gels containing SDS and double-stranded DNA, removal of SDS by washing of gels and staining of the gels with ethidium bromide.67) This method is simple and based on the principle that the association of DNA and DNA-binding proteins prevents the intercalation of ethidium bromide to DNA. We isolated the mt-nucleoids from several species of yeasts and detected candidates for the Abf2 homolog. A polyclonal antibody raised against S. cerevisiae Abf2 did not detect any mt-nucleoid proteins from other species, suggesting the diversity of Abf2 homologs. On the other hand, SDS-DNA PAGE detected at least a single protein among the mt-nucleoid proteins tested.68) We succeeded to detect and purify an Abf2 homolog of 17 kDa from Kluyveromyces lactis69) (Fig. 3), and an Abf2 homolog of 26 kDa from Pichia jadinii, which has a unique linear form mtDNA.70) Yeast species that are phylogenetically distant from S. cerevisiae had a tendency to have a molecular weight higher than 20 kDa. Taking advantage of the complete genome sequence, comparative analysis of the mt-nucleoid proteins from Klyuveromyces lactis and more phylogenetically diverse yeast species has been done.71) In these studies, the K. lactis Abf2 homolog displays only 28.5% amino acid sequence identity with Abf2 from S. cerevisiae, although the sequence identity of other mt-nucleoid proteins ranges between 50%–90%, suggesting the diversity of Abf2 homologs in the yeasts. Indeed, in mitochondria of phylogenetically distant yeast species like Schizosaccharomyces pombe or Yarrowia lipolytica, Abf2-like or putative mitochondrial HMG box proteins have not been identified by BLAST search.71) Therefore, we tried to isolate the mt-nucleoids from each species of yeast and identify the Abf2 homolog among the mt-nucleoid proteins.
There is a significant variety in size and form (circular or linear) of mtDNA molecules in yeasts.72),73) The yeast Candida parapsilosis has a 30-kbp linear form of mtDNA.74),75) We isolated the mt-nucleoids from C. parapsilosis and confirmed that the intact mtDNA is packaged without degradation in the isolated mt-nucleoids. The mt-nucleoids from C. parapsilosis contained a number of proteins including aconitase and a putative Abf2 homolog of 26 kDa. We purified the Abf2 homolog and identified a GCF1 gene that encodes the 26-kDa protein.76) In silico analysis of Gcf1 predicted the presence of a coiled-coil domain at the N-terminal region and a single HMG box at the C-terminal region using SMART (http://dylan.embl-heidelberg.de/) and COIL (http://www.ch.embnet.org/software/COIL_form.html), respectively (Fig. 3). Detection of only one HMG box by SMART analysis suggested that if a second HMG box were present, the amino acid sequence would be too weakly conserved to be discovered by in silico analysis.71) Our results indicated that Gcf1 is a novel type of mitochondrial HMG protein. The same type of HMG protein that retains a putative coiled-coil domain and one C-terminal HMG box has been identified in Candida albicans using in silico analysis.77) Divergence of amino acid sequences of the mt-nucleoid proteins was compared among S. cerevisiae and C. parapsilosis, and among C. albicans and C. parapsilosis, respectively. Interestingly, the results demonstrated that the HMG proteins like Abf2 and Gcf1 are the most diverged proteins among the mt-nucleoid proteins tested.76)
Subsequently, we isolated the mt-nucleoids from the yeast Yarrowia lipolytica, which is more phylogenetically distant from S. cerevisiae than C. parapsilosis.78) The yeast S. cerevisiae is able to grow by fermentation without mitochondrial respiration. This characteristic is different from human cells because almost all energy production of human cells depends on aerobic respiration of mitochondria. On the other hand, Y. lipolytica is an obligatory aerobic yeast whose growth exclusively depends on mitochondrial respiration. Therefore, Y. lipolytica is considered to be suitable as a model of human cells. We isolated the mt-nucleoids from stationary-phase cells and identified a 30-kDa DNA-binding protein, YlMhb1 (Y. lipolytica Mitochondrial HMG box-containing protein 1).78) Subsequently, we identified the YlMhb1 gene that encode this protein. Two putative HMG boxes were predicted using the Phyre server (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index), although the N-terminal HMG box domain was not detected by SMART (Fig. 3). This suggested that the N-terminal HMG box of YlMhb1 is weakly conserved. We confirmed that the Mhb1-GST fusion protein is targeted to the mt-nucleoids by fluorescence microscopy. The N-terminal HMG box (HMG1) and C-terminal HMG box (HMG2) were separately purified as GST-fusion proteins from E. coli. Our results demonstrated that the HMG1 box has higher affinity to double-stranded DNA than HMG2. This result suggested that HMG1 and HMG2 have different functions to maintain mtDNA. Deletion of the MHB1 gene decreased the copy number of mtDNA to 40% concomitant with the decrease of the number of mt-nucleoids, but did not alter the morphology of mitochondria. The expression of mitochondrial genes in Δmhb1 cells was markedly repressed, but respiration activity and the growth rate of Δmhb1 cells were indistinguishable from those of wild-type cells.78) These results suggest that unknown mechanisms compensate the defects in transcription of mitochondrial genes in Δmhb1 cells at the translational level.78)
Another defect in Δmhb1 cells is the increase of mutation rate of mitochondrial genes. First, the absence of YlMhb1 resulted in sensitivity to the intercalating agent, ethidium bromide. Second, a frequency of spontaneous oligomycin-resistant mutation in Δmhb1 cells is nearly 16-fold higher than that in wild-type cells. This frequency was further increased when cells were treated with MnCl2. These results indicate that YlMhb1 plays a role for protection of mtDNA against mutagenic events.78) YlMhb1 is predicted to serve in maintaining the copy number of mtDNA, facilitation of transcription of mitochondrial genes and suppression of mutation. On the other hand, the protein composition of the mt-nucleoids was not significantly altered in Δmhb1 cells, except for lack of Mhb1, as compared with SDS-PAGE.78)
Including human TFAM, S. cerevisiae Abf2, K. lactis Abf2, C. parapsilosis Gcf1 and Y. lipolytica Mhb1, comparison of mitochondrial HMG box proteins is shown in Fig. 3. Recently, Bakkaiova et al.79) compared the DNA-binding activity of Abf2, C. parapsilosis Gcf1 and Y. lipolytica Mhb1 with several types of DNA. They revealed that the region of the N-terminal HMG box 1 is also predicted in C. parapsilosis Gcf1 using the Phyre server. They found that the three HMG box proteins have different affinities to DNA. Abf2 and YlMhb1 bind quantitatively to double-stranded DNA with relatively low affinity, but CpGCf1 exhibited only negligible binding to double-stranded DNA.79) In contrast, they revealed that these proteins exhibit much higher affinity to recombination intermediates, such as Holliday junctions and replication forks, and suggested that the role of mitochondrial HMG box proteins is mediated by binding to recombination/replication intermediates.79)
We investigated how the size and number of the mt-nucleoids are affected by culture conditions of yeast cells80) (Fig. 4). The stationary-phase cells cultured in a modified Burkholder’s medium had on average 63 mt-nucleoids per cell, and each mt-nucleoid contained 3.9 copies of mtDNA. On the other hand, stationary-phase cells aerobically grown in the anaerobic (AN) medium, which is a nutrient-rich medium for anaerobic culture, contained on average 141 mt-nucleoids per cell, and each mt-nucleoid contained on average 1.5 copies of mtDNA.80) The most drastic changes of mt-nucleoid morphology were observed in anaerobic culture of cells. When cells were cultured to stationary phase in AN medium under nitrogen gas, highly aggregated mt-nucleoids containing 20 copies of mtDNA were formed (Fig. 4A). The average number of mt-nucleoids was 7 per cell. These observations indicated that the size and number of mt-nucleoids distinctly changes by response to the culture conditions of cells, but total mtDNA content of the cell does not markedly vary.
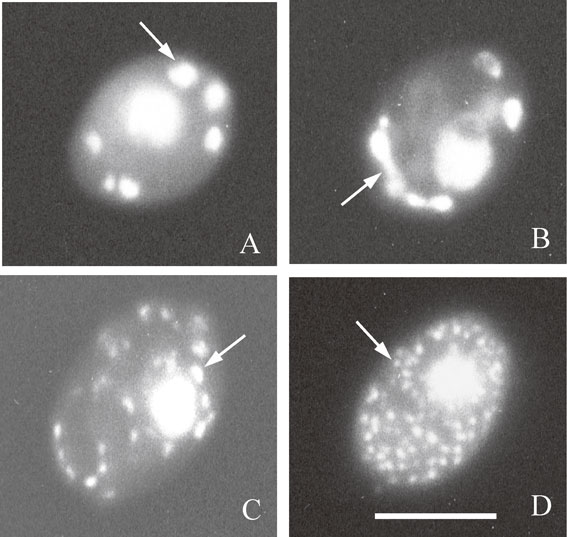
DAPI-fluorescence photomicrographs of four types of cells with different mt-nucleoids that appear during transfer of anaerobically grown cells to aerobic conditions. (A) An anaerobically grown cell that had largely aggregated mt-nucleoids at stationary phase. (B) A cell with elongated mt-nucleoids in the transition state at 2 h after transfer of anaerobically grown cells to aerobic conditions. (C) A cell with dispersed mt-nucleoids at 4 h after transfer. (D) A cell with dispersed small mt-nucleoids at 7 h after transfer. Arrows indicate the mt-nucleoids. Bar represents 5 µm. Figure 4 is from Ref. 81.
Distinct dynamic changes in the morphology of mt-nucleoids occur by transfer of anaerobically grown cells to aerobic culture (Fig. 4). In anaerobic conditions, yeast cells synthesize ATP by alcohol fermentation, and respiration activity was markedly decreased. When anaerobically grown cells that had large aggregated mt-nucleoids (Fig. 4A) were transferred to aerobic conditions, the aggregated mt-nucleoids rapidly dispersed to small particles during 7 hours, concomitant with rapid increase of respiration activity (Fig. 4BCD).81) The highly aggregated mt-nucleoids could be isolated from anaerobically grown cells with sucrose gradient centrifugations.82) This indicated that the highly aggregated mt-nucleoids observed by DAPI staining are not the apparent assembly of small mt-nucleoids.
We recently compared the morphology of mitochondria and mt-nucleoids between rho+ cells and rho− cells.83) We found that major forms of mitochondria are tubular network in log-phase cells regardless of respiration activity. However, the morphology of mt-nucleoids differed among strains. Wild-type rho+ cells and rho− cells with a long unit length of mtDNA sequence had the mt-nucleoids with strings-of-bead appearance in tubular mitochondria. However, the mt-nucleoids in rho− cells with multiple repeats of the short ori5 sequence appeared as large aggregates. As yeast mtDNA performs multiple recombinations at homologous regions of mtDNA, we speculate that mtDNA with short repeats of mtDNA sequence is interconnected by homologous recombination to form aggregated large mt-nucleoids in rho− cells.
Several lines of evidence show that mitochondria of S. cerevisiae are transmitted to the bud from mother cells by interaction of the outer surface of mitochondria with actin filaments. First, it was found that many amino acid substitutions of yeast actin markedly affected the mitochondrial organization.84) Second, rhodamine-labeled F-actin was shown to slide in vitro along the surface of mitochondria that were fixed on a glass slide in an ATP-dependent manner.85) On the other hand, strains bearing mutations of each myosin (I to V) exhibited normal mitochondrial movement from mother cell to bud (anterograde movement).85) Therefore, at that time, it was proposed that myosin does not serve as a force generator for mitochondrial movement, and instead, actin polymerization and the Arp2/3 complex serve as force generators for actin-dependent, anterograde mitochondrial movement.86) In contrast, it has been reported that conditional myo2 mutants encoding a class V myosin have defects in mitochondrial movement towards the bud.87)–89) Current evidence supports the model that the function of Myo2 is essential for mitochondrial inheritance and requires either Mmr1 or Ypt11.90) The mt-nucleoids are attached to a part of the inner membrane, and transmitted to the bud with transmission of mitochondria. The image of the interaction of mitochondria, actin cable and the plasma membrane is illustrated in Fig. 6B.
The distribution of tubular mitochondria in the fission yeast, Schizosaccharomyces pombe, is mediated by interaction of mitochondria and microtubules.91) A recent study demonstrated that mitochondria of S. pombe are distributed passively through attachment to dynamic microtubules. Kinesins and dynein play a negligible role in mitochondrial distribution of S. pombe.92) As far as we observed in different yeasts, presence of tubular mitochondria is a general feature in yeast cells. In the unique triangular yeast, Trigonopsis variabilis, tubular mitochondria also highly developed and transmitted to the bud.93) The budding yeast Saccharomycodes ludwigii and Yarrowia lipolytica also have tubular mitochondria.94),95) It may be possible that mitochondrial distribution is regulated in either actin-dependent or tubulin-dependent manners in each yeast species.
Mitochondria and the mt-nucleoids can be visualized in both living cells and chemically fixed cells, taking advantage of mitochondria-targeted GFP (mtGFP) or mtRFP and DAPI, respectively96) (Fig. 5). In log-phase cells, the mt-nucleoids are aligned like strings-of-beads in tubular mitochondria elongating to the bud from the mother cell (Fig. 5ABC). Although tubular mitochondria are dominant in log-phase cells, mitochondria frequently fuse and divide in those cells. On the other hand, in stationary-phase cells, tubular mitochondria divide into small ones and are dispersed in the cytoplasm (Fig. 5DEF).
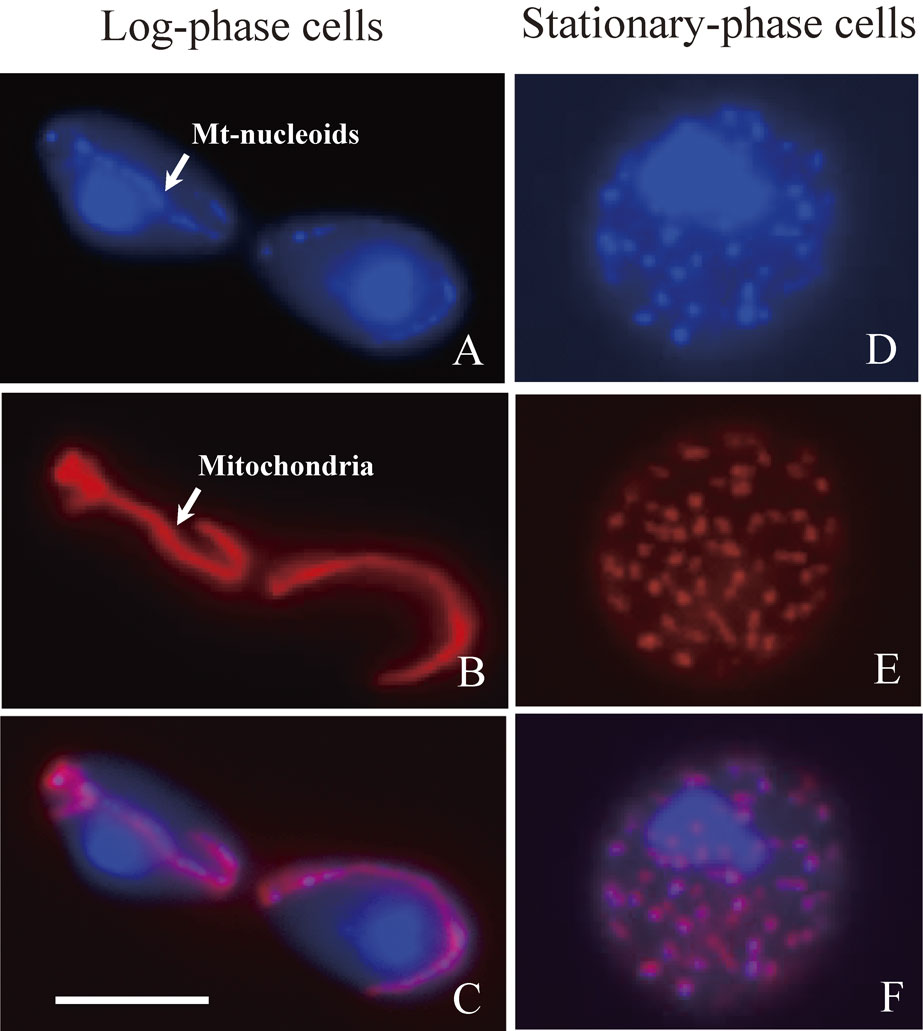
Morphology of mitochondrial nucleoids and mitochondria in log-phase and stationary-phase cells. Mitochondria are labeled with mitochondria-targeted RFP (mtRFP) by transformation of cells with a plasmid pVT100U-mtRFP.96) Cells at log phase or stationary phase were fixed with 3.7% formaldehyde for 1 h at room temperature. After washing cells with NS buffer,16) cell nuclei and mt-nucleoids were stained with DAPI. (A and D) Cell nuclei and mt-nucleoids stained with DAPI. (B and E) Mitochondria labeled with mtRFP. (C and F) Merged images of A and B, D and E, respectively. In log-phase cells, the mt-nucleoids look like strings-of-beads in tubular mitochondria (arrows in Fig. 5A and Fig. 5B). In stationary-phase cells, the mt-nucleoids are maintained in small spherical mitochondria. Bar represents 5 µm.
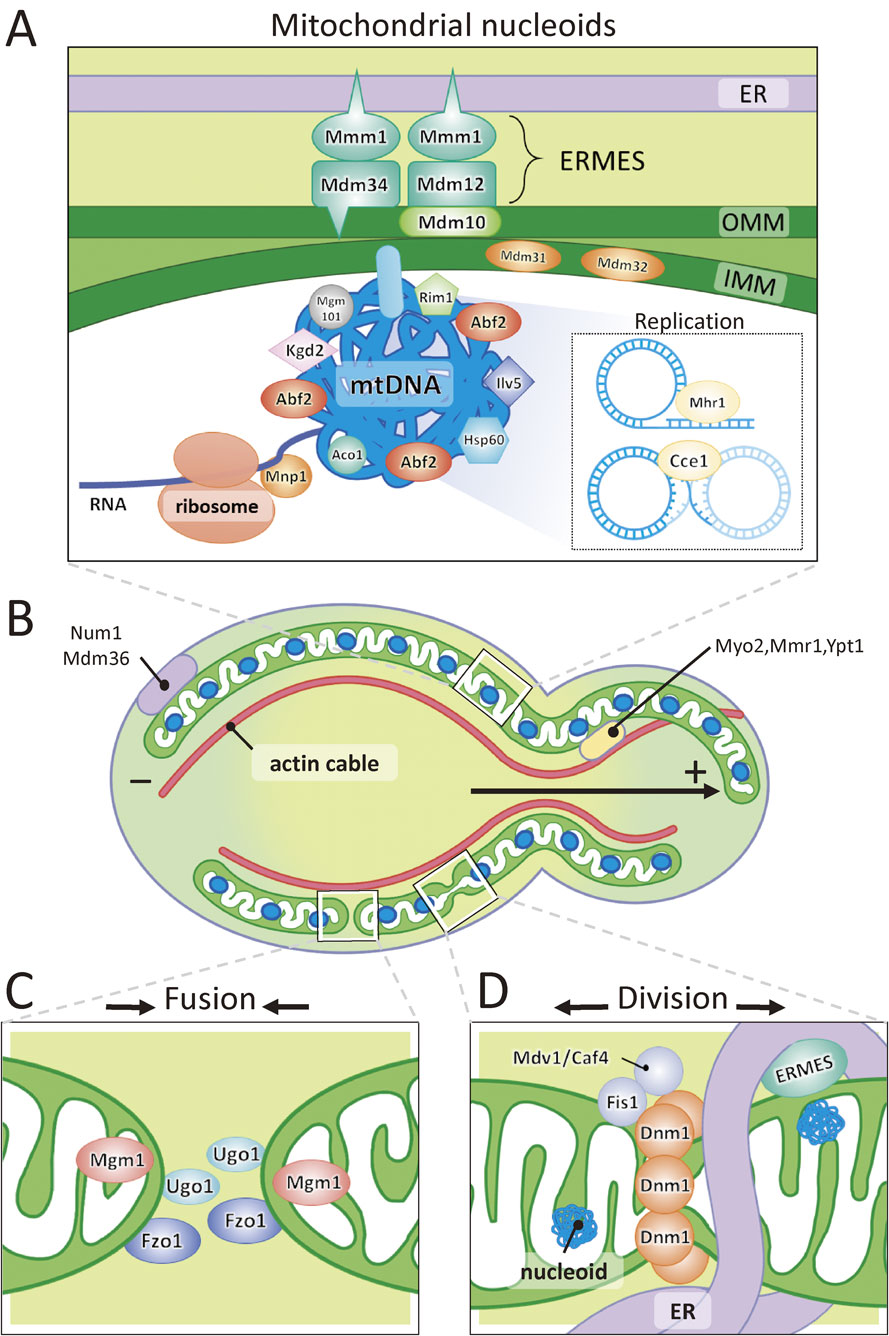
A model for the organization of mt-nucleoids, transport of mitochondria, and mitochondrial fusion and division. (A) The mtDNA is compactly packaged in the mt-nucleoids by association with Abf2 and other proteins. The mt-nucleoids are located in the vicinity of ERMES, where the ER contacts with the outer and inner mitochondrial membrane. The mt-nucleoids are attached to a part of the inner membrane. Rolling-circle replication and recombination of mtDNA occur in the mt-nucleoids. (B) Maintenance of the shape of mitochondria and transport of tubular mitochondria are performed by interaction with actin cables and the plasma membrane. (C) Fusion of mitochondria is mediated by Fzo1, Ugo1 and Mgm1. (D) Division of mitochondria is carried out by recruitment of cytoplasmic Dnm1 by Fis1 and Mdv1/Caf4. Recent findings demonstrate that mitochondrial fission sites are determined by the ER-mitochondria encounter structure (ERMES).104)
Since the discovery of plasticity of mitochondrial morphology, researchers have focused on how mitochondrial morphology is regulated. To this end, many mutants that exhibit abnormal mitochondrial morphology were screened by means of mutagenesis and fluorescence microscopic observation by staining of mitochondria with 2-(4-dimethylaminostyryl)-1-methylpyridinium iodide (DASMPI), DiOC6(3) and MitoTracker, or by labeling of mitochondria with mtGFP. Using this strategy, many genes that participate in the regulation of mitochondrial morphology were identified. Among them, a dnm1 mutant that had a defect in division of mitochondria was isolated.97),98) Moreover, an fzo1 mutant was isolated that exhibits aberrant mitochondrial morphology similar to fuzzy onion in the spermatogenesis of Drosophila melanogaster.99) FZO1 encoded a mitochondrial GTPase that is located on mitochondrial outer membrane. The yeast homolog of FZO1 was soon identified and the fzol mutant of S. cerevisiae also displayed defects in mitochondrial fusion.100),101)
Mitochondrial fusion requires mitochondrial outer membrane proteins, Fzo1, Ugo1 and the inner membrane protein Mgm1. Fzo1 and Mgm1 have a GTPase activity and are required for fusion of the outer membrane and inner membrane, respectively (Fig. 6C). On the other hand, mitochondrial division involved three components of Dnm1, Fis1 and Mdv1/Caf4. Dnm1p has GTPase activity and serves as division machinery that mediates mitochondrial scission. Dnm1 is a cytosolic protein recruited to the division site by Fis1 and Mdv1/Caf4, which are outer membrane proteins (Fig. 6D). Accordingly, the mitochondrial morphology of yeast is regulated by a balance of fusion and division. The Δdnm1 cells have a highly complicated network of mitochondria and are able to maintain normal mtDNA. In contrast, the Δfzo1 cells are devoid of mtDNA (rho0). These results suggest that mitochondrial fusion is essential for maintenance and normal inheritance of mt-nucleoids for unknown reasons. Additionally, Num1 functions to tether the cortical network of mitochondria to the plasma membrane102) (Fig. 6C).
Recent research revealed that the interactions of the ER (endoplasmic reticulum) and mitochondria also have important roles in determining mitochondrial division sites. The structure that tethers the ER and mitochondria is called ERMES (ER-Mitochondria Encounter Structure)103),104) (Fig. 6AD). The core components of ERMES are at least composed of four proteins: Mdm10, Mdm34, which are integral outer mitochondrial membrane proteins, Mmm1, which is an ER membrane protein and Mdm12, which is a cytosolic protein. ERMES determines the site for ER-mediated mitochondrial division sites and constricts mitochondria. Following these steps, Dnm1 is recruited and assembled to the division site. Interestingly, ERMES is assembled nearby the mt-nucleoids104) (Fig. 6D). When division of mitochondria occurs, mt-nucleoids also divide and are frequently distributed to both tips of the separated mitochondria.104) Mitochondrial division without mt-nucleoid division also occurs. In that case, the mt-nucleoids were distributed to the single tip of divided mitochondria. This mechanism ensures that each mitochondrion has its own genome. It still remains to be determined how ERMES located on the outer mitochondrial membrane recognizes the position of mt-nucleoids located in the matrix. Actively replicating mt-nucleoids are also linked to ER-mitochondria contacts in human cells.105) A predicted image of a mitochondrial division site is illustrated in Fig. 6D.
During the course of observation of mt-nucleoids and mitochondria, we found the presence of mitochondrial inclusion bodies in large mitochondria of yeast cells, which emitted blue fluorescence under UV excitation. We isolated mitochondria that included fluorescent inclusion bodies and then identified the major component of mitochondrial fluorescent inclusion body as a mitochondrial aldehyde dehydrogenase, Ald4.106) Subsequently, we confirmed the localization of Ald4 in the inclusion body by means of both immunoelectron microscopy and immunofluorescence microscopy.106) Mitochondria with mitochondrial inclusion bodies are very long in length and elongate straightforward. A very small number of mt-nucleoids are distributed in mitochondria with mitochondrial inclusion bodies (unpublished data). Although mitochondrial morphology is supposed to be determined by interaction of mitochondria and the structure outside mitochondria, it is also true that the mitochondrial morphology is determined by intra-mitochondrial structures in this particular case.
Although the unit of mtDNA in S. cerevisiae is 75–85 kbp circular DNA, long concatemeric molecules have been detected by pulsed field gel electrophoresis in both the yeasts Torulopsis glabrata and S. cerevisiae.107) Meleszka et al.107) predicted rolling-circle replication of yeast mtDNA. Subsequently, Ling and Shibata108) demonstrated that an mhr1-1 mutation exhibits a defect in the partitioning of nascent mtDNA into buds. Mhr1 protein (Mhr1) has activity to pair single-stranded DNA and homologous double-stranded DNA to form heteroduplex joints in vivo. As incorporation of deoxybromouridine in newly synthesized mtDNA occurred in mitochondria located near the budding site in mother cells, Ling and Shibata108) suggested that mtDNA replication take place by a rolling-circle mechanism, which is mediated by Mhr1p near the budding site, and newly synthesized mtDNA is transmitted to the bud. They also provided evidence to show that rolling-circle replication and transmission of a few selected mtDNA to the bud is a possible mechanism to establish homoplasmy109) (Fig. 6A). Cce1 is a cruciform cutting enzyme that functions in resolution of the recombination junction of mtDNA (Fig. 6A). The deletion of CCE1 gene resulted in large aggregates of mt-nucleoids.110) This indicates that recombination of mtDNA affects the morphology of the mt-nucleoids. Deletion of ABF2 gene resulted in defects of transmission of mt-nucleoids.31) This suggests that Abf2 is also involved in the recombination and transmission of mtDNA at replication sites in the mother cells. Although both Abf2 and Cce1 are involved in the recombination of mtDNA, recent study demonstrated that Abf2 does not significantly affect either binding of Cce1 to branched DNA or rate and specificity of Holiday junction resolution.111)
There are still arguments about the replication mechanism of yeast mtDNA. Gerhold et al.112) showed that, although Candida albicans has a circular genetic map, C. albicans mtDNA forms a complex branched network that does not contain detectable amounts of circular molecules. These results suggested that the recombination-driven replication rather than rolling-circle replication is the major or only means of mtDNA replication.113) Gerland et al.114) also reported that the linear mtDNA with telomeres also employs recombination for replication initiation in Candida parapsilosis.
The unicellular red alga, Cyanidioschyzon merolae has one nucleus, one mitochondrion and one chloroplast.115) Each organelle divides once during the cell cycle. The timing of mitochondrial DNA replication, division of mt-nucleoids and division of mitochondria is strictly regulated to coordinate these events with the cell cycle. In contrast, in the yeast S. cerevisiae, the replication of mtDNA continuously proceeds throughout the cell cycle. Furthermore, mitochondria are organized in a network of tubular structures, which frequently divide and fuse with each other through vegetative growth.
A lot of knowledge has been accumulated on the organization of mt-nucleoids and mitochondrial dynamics in yeast cells. However, several questions remain to be answered. The mechanisms of hypersuppressiveness and segregation of mtDNA from heteroplasmy to homoplasmy may be explained by selective replication of mtDNA via rolling-circle mechanisms. On the other hand, the relation between morphological changes of the mt-nucleoids and rolling-circle replication of mtDNA is ambiguous.
Abf2 was proven to be an important protein that functions in stable inheritance of mtDNA. However, overexpression of Abf2p leads to severe destabilization of mtDNA. How do mitochondrial HMG box proteins contribute to the stable maintenance of mtDNA, except for the role of packaging of mtDNA? Why are the amino acid sequences of HMG box proteins most diverged among mt-nucleoid proteins? This is in contrast to nuclear histone, a highly conserved protein that plays an essential role for packaging of DNA in eukaryotes. In yeasts, there is significant diversity among the forms of the mtDNA (circular DNA or linear DNA) and their genome size. Are the significant changes in nucleotide sequence and form of mtDNA related to the diversity of HMG box proteins? As listed in Table 1, there are several bi-functional mt-nucleoid proteins that serve for stability of mtDNA, except for Abf2. How do these bi-functional proteins link the metabolic activity of mitochondria to the expression of mitochondrial genes?
The yeast S. cerevisiae has mitochondria that are different from human mitochondria because respiratory-deficient rho− cells with deleted mtDNA can be easily produced and these cells can survive on fermentative substrates. From this viewpoint, mitochondria of obligate aerobic yeast Y. lipolytica can serve as a model of human mitochondria. The mt-nucleoids play an essential role for protecting mtDNA from oxidative stress derived from respiration. Mutation of the mt-nucleoid proteins will result in damage or loss of mtDNA, which is related to human diseases.116) Therefore, research on yeast mt-nucleoids will continue to contribute to the fundamental understanding of the function of mt-nucleoids.

Isamu Miyakawa was born in Toyama Prefecture in 1955. He graduated from Tohoku University in 1977 and in the same year entered the Graduate School of Sciences, Tohoku University. He received his Ph.D. degree from Tohoku University in 1986. He started his research career as an Assistant Professor in the Department of Biology, Faculty of Sciences, Yamaguchi University in 1980. He was promoted to an Associate Professor in 1994 and to a Professor in 2003. He is currently working as a Professor in Department of Biology, Faculty of Sciences, Yamaguchi University (Department of Biology, College of Sciences, Graduate School of Sciences and Technology for Innovation, Yamaguchi University). He studied the contraction mechanisms of Ascidian smooth muscle under the direction of Prof. Kazuhiko Konishi (1976–1979). From 1980, he started his research on the morphology of yeast mitochondria and the mitochondrial nucleoids with Prof. Nobundo Sando, Yamaguchi University and Prof. Tsuneyoshi Kuroiwa, National Institute for Basic Biology. He spent 10 months in 1990 at Curie Institute, Orsay, France, where he studied the diversity of yeast mitochondrial genome in Hiroshi Fukuhara’s laboratory. He has found the dynamic morphological changes in yeast mitochondria and the mitochondrial nucleoids during the life cycle of yeast. He has contributed to the studies on the organization and dynamics of yeast mitochondrial nucleoids.
We thank Prof. Tsuneyoshi Kuroiwa and Prof. Nobundo Sando for their continuous support. This research was partly supported by JSCP KAKENHI Grant Number 15K07168, by the Core Research for Evolution Science and Technology (CREST) Program of Japan Science and Technology Agency (JST), and by MEXT TOKUBETSUKEIHI to Isamu Miyakawa, Yamaguchi University. We thank Ms. Hana Taura for the illustrations of figures.