2022 Volume 98 Issue 8 Pages 470-492
2022 Volume 98 Issue 8 Pages 470-492
Land plants have developed sophisticated systems to cope with severe stressful environmental conditions during evolution. Plants have complex molecular systems to respond and adapt to abiotic stress, including drought, cold, and heat stress. Since 1989, we have been working to understand the complex molecular mechanisms of plant responses to severe environmental stress conditions based on functional genomics approaches with Arabidopsis thaliana as a model plant. We focused on the function of drought-inducible genes and the regulation of their stress-inducible transcription, perception and cellular signal transduction of stress signals to describe plant stress responses and adaptation at the molecular and cellular levels. We have identified key genes and factors in the regulation of complex responses and tolerance of plants in response to dehydration and temperature stresses. In this review article, we describe our 30-year experience in research and development based on functional genomics to understand sophisticated systems in plant response and adaptation to environmental stress conditions.
Plants are sessile organisms and are able to adapt to severe environmental conditions for growth, development and survival. During evolution in stressful environmental conditions, land plants have developed unique systems to propagate environmental stress signals from sensing tissues to appropriate targets to achieve optimal growth maintenance and tolerance to stress conditions.1),2) Land plants respond and adapt to adverse environmental conditions, such as drought, high and low temperatures, and high salinity. During drought stress, plants close stomata to save water, and photosynthesis is prohibited by the reduction in gas exchange. Abscisic acid (ABA) is a key phytohormone that not only prevents water loss by transpiration from guard cells but also enables adaptation to drought stress conditions. ABA accumulates in drought stress conditions and quickly spreads intercellularly in whole tissues to induce plant stress responses and adaptation.3)–6) In addition to physiological research, molecular and cellular biology have contributed to resolving the precise regulatory gene networks associated with complex abiotic stress responses and resistance in model plants such as Arabidopsis thaliana and rice in abiotic stress conditions.4),7),8)
When we started our laboratory at the RIKEN Tsukuba Life Science Center in 1989, we focused our work on molecular biological analyses of plant environmental responses, especially drought stress, because water deficit conditions have a severe effect on plant growth. We selected Arabidopsis thaliana as a model plant for this purpose because of its high potential for genomics research.9) In the late 1980s, protein analyses revealed the accumulation of specific proteins in response to low and high temperature and water deficiency. Accumulated proteins include late embryogenesis abundant (LEA) proteins and heat shock proteins, which function in protecting cells from stress conditions.1) These observations led us to hypothesize stress-inducible expression of stress-specific genes whose products function in stress tolerance and response in complex environmental stresses.9) To understand the molecular and cellular basis of plant responses to complex abiotic stress, especially drought, cold, and heat stresses, we have used reverse genetics and functional genomics approaches to isolate many drought-inducible genes with various functions and analyze their regulatory systems of gene expression in plant abiotic stress responses.4),10) We focused our work on analyzing transcriptional regulation in abiotic stress responses and their associated signaling networks (Fig. 1A). We discovered many important regulatory genes involved in plant responses to drought, cold, and heat and analyzed gene expression and signal transduction in abiotic stress responses. We discovered ABA-independent regulatory systems in the plant response to drought stress in addition to those that are ABA dependent.10) We also analyzed ABA-dependent regulatory processes of drought stress responses, including transcriptional regulation, signal transduction and intertissue communication.7),11) Moreover, we have tried to apply basic knowledge obtained with Arabidopsis for application to the breeding of drought-tolerant crops.7),12)

Plant responses to drought stress: From regulatory gene networks in cellular responses to integrative interorgan signaling for stress tolerance. A. Molecular genetics and functional genomics have elucidated the complex cellular processes of plant responses to environmental stresses, including regulation of gene expression and cellular signal transduction. B. Plant hormones, including ABA and peptides, function in intertissue and interorgan signal transduction for the integration of stress signals driving proper responses throughout the entire plant. This figure is reprinted with modification from Ref. 17.
In this review article, we mainly focus on our functional genomics studies to understand complex plant responses to abiotic stress, especially drought and temperature stresses, and regulatory systems for abiotic-stress tolerance.7),11),13),14) Recent studies on intertissue and interorgan signaling in plant stress responses and adaptation are also summarized (Fig. 1B).15)–17) Moreover, the importance of quantitative phenotyping analyses of plant growth in water-deficit conditions is discussed.17) The application of basic knowledge on plant stress resistance for the molecular breeding of drought-tolerant crops is discussed to contribute to crop production in dry areas during global warming.12) In this review, we summarized our research strategies and accomplishments on molecular and cellular analyses of regulatory systems of plant responses to environmental abiotic stresses, such as drought, cold, and heat stresses.
Since 1989, we have cloned numerous genes involved in the environmental stress response and resistance acquisition, analyzed their gene structure and function, and then analyzed the regulation of gene expression in drought and low-temperature conditions. First, we prepared cDNA libraries from Arabidopsis plants that were dehydrated and screened cDNAs of the mRNAs that were upregulated in response to drought stress.18),19) We have shown that many genes are induced by abiotic stress, such as drought and low temperature, and that many of these genes are involved in the acquisition of stress tolerance. Later, we used microarrays to identify drought-upregulated genes in Arabidopsis and other crops.20),21) Drought stress-inducible genes at the transcriptional level include not only genes regulated by the plant hormone ABA but also genes induced by stress without ABA. ABA is produced in water-deficit conditions and plays an important role in plant drought tolerance. Exogenous application of ABA also induces several genes that respond to dehydration.22) Several reports have described genes that are induced by dehydration and cold stresses but that do not respond to exogenous ABA treatment.22)–24) This suggests the existence of ABA-independent, as well as ABA-dependent, signal transduction cascades between the initial stress signals and the expression of specific stress-related genes. Transcriptional regulation plays important roles in the acquisition of the environmental stress response and tolerance in plants.
The expression of many plant genes with various functions is regulated by abiotic environmental stresses such as drought, heat, and cold. Transcriptome analysis using microarray technology is a powerful technique that has proven to be very useful for the discovery of many stress-inducible genes involved in stress response and tolerance.20),21) Numerous genes that are induced by various abiotic stresses have been identified using different microarray systems.10),25)
Genes induced during stress conditions are thought to function not only in protecting cells from stress by the production of important metabolic proteins but also in the regulation of genes for signal transduction in the stress response (Fig. 2). Thus, these gene products are classified into two major groups with different functions.9),10) The first group includes proteins that probably function in stress tolerance, such as chaperones, late embryogenesis abundant (LEA) proteins, osmotin, antifreeze proteins, key enzymes for osmolyte biosynthesis such as proline and raffinose, water channel proteins, sugar and proline transporters, detoxification enzymes to reduce reactive oxgen spieces (ROS), enzymes for fatty acid metabolism, and proteinase inhibitors. Overexpression of these stress-inducible genes that encode key enzymes for osmolyte biosynthesis, LEA proteins, and detoxification enzymes has been shown to improve stress-tolerant phenotypes of transgenic plants.12),25) These results indicate that the gene products of the stress-inducible genes truly function in stress tolerance in plants.

Functions of drought-inducible genes in stress tolerance and responses. Thousands of drought-inducible genes have been identified by transcript analyses. These gene products are classified into two major groups: functional proteins involved in cellular stress tolerance and regulatory proteins involved in gene expression and signal transduction of stress responses. In addition to these proteins, noncoding RNAs and small peptides have been reported to function in the regulation of stress responses. Names and functions of genes for stress-inducible genes are summarized in Table 1.
The second group contained protein factors involved in the further regulation of signal transduction and gene expression that probably function in the stress response (Fig. 2). They include various types of transcription factors, suggesting that various transcriptional regulatory mechanisms function in drought-, cold- or high salinity-stress signal transduction pathways.9)–11) These stress-inducible transcription factors include members of the DRE-binding protein (DREB), ethylene-responsible element binding factor (ERF), zinc-finger, WRKY, MYB, basic helix-loop-helix (bHLH), basic-domain leucine zipper (bZIP), NAC, and homeodomain transcription factor families.10) These transcription factors can cooperatively regulate various stress-inducible genes and may constitute transcriptional gene networks. Functional analysis of these stress-inducible transcription factors should provide more information on the complex regulatory gene networks that are involved in responses to drought, cold, and high-salinity stresses. The others were protein kinases, protein phosphatases, enzymes involved in phospholipid metabolism, and other signaling molecules. Some of these stress-inducible regulatory genes have been overexpressed in transgenic plants to generate stress-tolerant phenotypes in transgenic plants.2),7),12)
Many noncoding RNA transcripts have also been identified as stress-inducible genes. These include microRNAs, antisense RNAs and long-noncoding RNAs.26),27) Some of these noncoding RNAs function in the regulation of stress responses. Stress-inducible expression of regulatory genes is a typical response and adaptation of plants against adverse environmental stress conditions.
Table 1 summarizes a list of names of genes and factors that we discovered during our research on plant responses to abiotic stress like drought, heat, and cold.
| Gene name | Functions | Abiotic stress | References |
|---|---|---|---|
| Stress-inducible genes involved in stress tolerance | |||
| LEA | Late embryogenesis abundant | Dry, Cold | 10 |
| RD29 | Responsive to dehydration 29 | Dry, Cold, ABA | 10, 18 |
| RD22 | Responsive to dehydration 22 Unidentified seed protein |
Dry, ABA | 10, 22 |
| ERD1 | Early responsive to dehydration 1 Clp protease regulatory subunit |
Dry, ABA | 10, 19, 42 |
| HSP | Heat shock protein | Heat, Dry | 10, 13 |
| GolS | Galactinol synthase | Dry, Cold, Heat | 61–63 |
| GAD | GABA decarboxylase | Dry, ABA | 61 |
| Transcription factors | |||
| DREB1/CBF | DRE-binding protein 1/ C-repeat binding factor |
Cold | 10, 28, 30 |
| DREB2 | DRE-binding protein 2 | Dry, Heat | 10, 28, 34 |
| AREB/ABF | ABRE-binding protein TF | ABA, Dry | 10, 36, 37 |
| AtMYC2 | MYC related TF 2 | ABA, Dry | 10, 39, 40 |
| AtMYB2 | MYB related TF 2 | ABA, Dry | 10, 40 |
| NAC/RD26 | NAC TF | Dry | 10, 43, 44 |
| ZF-HD | Zn finger-homeodomain TF | Dry | 10, 45 |
| HB6 | Homeobox 6 TF | ABA, Dry | 10, 46 |
| TCP13 | TCP family 13 TF | Dry | 47 |
| bZIP60 | bZIP TF in ER stress response | ER | 48 |
| bZIP17/28 | bZIP TF in ER stress response | ER | 49 |
| TAF12b | TATA binding factor 12b | ER | 50 |
| HSFA1 | Heat shock factor A1 | Heat | 13 |
| CAMTA1 | Calmodulin-binding TF | Cold | 14, 56, 57 |
| RVE | Reveille TF | Cold | 14, 57 |
| ABA related enzymes and transporters | |||
| NCED | 9-cis-epoxycarotenoid dioxygenase | ABA synthesis | 104–106 |
| CYP707A | P450 cytochrome oxidase | ABA degradation | 104–106 |
| ABC | ATP-binding cassette transporter | ABA transport | 110–114 |
| NPF | Nitrate/peptide transporter | ABA transport | 116–118 |
| DTX | Detoxification efflux carrier | ABA transport | 119 |
| Signaling factors in stress responses | |||
| SnRK2 | SNF-like protein kinase 2 | ABA, Dry | 11, 67, 68 |
| PP2C | Protein phosphatase 2C | ABA, Dry | 3, 5 |
| PYR/PYL/RCAR | START-type ABA receptor | ABA | 73, 74 |
| Raf-like kinase | MAPK kinase kinase (MAPKKK) | ABA, Dry | 81–83 |
| VCS | VARICOSE mRNA decapping | Dry | 11, 84 |
| CK1 | Casein kinase 1 | Dry | 55 |
| CPK | Ca2+-dependent protein kinase | Dry, Cold, ABA | 11 |
| ATHK1/AHK1 | Arabidopsis histidine kinase 1 | Dry | 87–89 |
| MCA1 | MID1 homolog of yeast Ca2+-permeable stretch-activated channel |
Dry, Cold | 92, 93 |
| OSCA1 | Hyperosmolarity induced Ca2+ increase | Dry | 96–98 |
| HPCA | H2O2-induced Ca2+ increase | ROS | 101 |
| RBOHD | Respiratory burst oxidase D | ROS | 101, 103 |
| CLE25 | Clavata3 related peptide 25 | Dry | 121 |
Genes and factors involved in abiotic stress responses and tolerance are listed in this table and discussed in the text.
Dry: drought stress; Cold: cold stress; Heat: heat stress; ABA: abscisic acid regulated; ER: endoplasmic reticulum; ROS: reactive oxygen species; TF: transcription factor.
The expression patterns of genes induced by abiotic stresses have been analyzed by RNA-blot hybridization analyses and recently using microarrays, RNA sequencing, and quantitative PCR. Through these transcriptome studies, thousands of plant genes have been shown to be stress inducible. Only 10% of drought-inducible genes were also induced by cold stress.21) The results also indicated broad variations in the timing of induction of these genes, and there are at least two groups showing different expression profiles.17) In one group, gene expression was rapid and transient in response to drought and cold stresses, reached a maximum within several hours, and then decreased. Most of these genes encoded regulatory protein factors such as bHLH transcription factors, DREB1A, DREB2A, AP2/ERF domain-containing protein RAP2, and growth factor-like protein.10) In the other group, gene expression slowly and gradually increased after stress treatment within 10 hours. Most of these genes encode functional proteins such as LEA proteins, detoxification enzymes and enzymes for osmolyte synthesis. However, some of these stress-inducible genes respond to ABA, whereas others do not. ABA-deficient mutants were used to analyze drought-inducible genes that respond to ABA. Many genes were induced by exogenous ABA treatment. However, some genes were induced by cold or drought in ABA-deficient (aba) or ABA-insensitive (abi) mutants. This means that there are not only ABA-dependent systems but also ABA-independent systems involved in the drought stress response (Fig. 3).10)

Transcriptional regulatory gene network in plant abiotic stress responses, such as drought, heat, and cold stresses. ABA is a major plant hormone associated with the drought stress response. There are ABA-dependent regulatory systems and ABA-independent systems involved in the drought stress response. The bZIP transcription factors AREBs/ABFs function in ABA-dependent regulation. In ABA-independent pathways, DREB2s encoding AP2 transcription factor function in drought-inducible transcription. CK1 protein kinase is involved in the activation of DREB2s. In addition, NAC, ZF-HD, MYB, and MYC transcription factors are involved in the drought stress response. In the heat stress response, HSFA2 transcription factors are major regulatory factors and regulate downstream transcription factors, including DREB2s. In the cold stress response, DREB1s/CBFs are major transcription factors in cold-inducible transcription. Cold-inducible induction of DREB1/CBF genes is regulated by 2 different transcription factors: CAMTA TF in the rapid response and CCA1/LHY and RVE in circadian regulation. Names and functions of genes for transcription factors (TFs; shown in circles) and signaling factors (SFs; shown in hexagons) are summarized in Table 1. Cis-acting elements are shown as rectangles. Receptors and sensors have not been clearly identified and shown with dashed lines.
Transcription factors involved in ABA-dependent and ABA-independent gene expression in abiotic stress responses are summarized in Table 1.
We precisely analyzed the expression profiles of the Arabidopsis RD29A/COR78/LTI78 gene, the product of which protects cells from dehydration. The RD29A gene is induced by drought, cold, and ABA treatment. However, this gene is induced in aba or abi mutants by both drought and cold stresses, which indicates that it is governed by both ABA-dependent and -independent regulation in drought and cold conditions.22) Analysis of this RD29A promoter has shown that a 9-bp conserved sequence, TACCGACAT, named the dehydration responsive element (DRE), is an essential cis-acting element for the regulation of RD29A induction in the ABA-independent response to dehydration and cold.28) DRE is also found in the promoter regions of many cold-inducible genes and is named C-repeat (CRT) and low-temperature-responsive element, both containing an A/GCCGAC motif that forms the core of the DRE sequence and regulates cold-inducible promoters.29) The cDNAs encoding DRE/CRT binding proteins, C-repeat binding factor/DRE binding protein 1 (CBF/DREB1), and DREB2, have been isolated using yeast one-hybrid screening.30),31) These proteins contained the conserved DNA-binding domain found in the ethylene-responsive element binding factor (ERF) and Apetala2 (AP2) proteins. These proteins specifically bind to the DRE/CRT sequence and activate the transcription of genes driven by the DRE/CRT sequence (Fig. 3).
In Arabidopsis, three genes encoding DREB1/CBF lie in tandem on chromosome 4 in the following order: DREB1B/CBF1, DREB1A/CBF3, and DREB1C/CBF2. Expression of the DREB1/CBF genes is induced by cold stress but not by dehydration or high-salinity stress. Three DREB1 proteins are major transcription factors involved in cold-induced gene expression.30)–33) There are two DREB2 proteins, DREB2A and DREB2B. Expression of the DREB2 genes is induced by dehydration and high-salinity stresses but not by cold stress.34) Both DREB2A and DREB2B proteins function in drought responses and are discussed in more detail in a later section on roles of DREB2.
ABA plays an important role in the signal transduction of osmotic stress in plants. ABA-responsive element (ABRE: ACGTGG/TC) is a major cis-acting element that functions to regulate ABA-responsive gene expression.10),35) In Arabidopsis, two ABRE motifs are important for the regulation of ABA-responsive expression of the RD29B gene encoding a LEA-like protein. The basic leucine zipper (bZIP) transcription factor ABRE-binding proteins (AREB)/ABRE-binding factors (ABF) bind to ABREs and activate ABA-dependent gene expression.36) Activation of the AREB1/ABF2 (AREB1) and AREB2/ABF4 (AREB2) proteins requires ABA-mediated posttranslational phosphorylation. ABA-activated protein kinases, including SNF1-related protein kinase 2 (SnRK2), phosphorylate and activate AREB/ABF-type proteins.37),38) The perception of ABA and activation of AREB/ABFs is discussed in a later section on ABA signaling.
The induction of the drought-inducible Arabidopsis gene RD22, which encodes a protein with homology to an unidentified seed protein, is mediated by ABA. A MYC transcription factor, AtMYC2, and a MYB transcription factor, AtMYB2, bind the RD22 promoter and cooperatively activate RD22.39) These transcription factors are synthesized after endogenous levels of ABA accumulate, which indicates their role in a late stage of the drought response. Overexpression of both AtMYC2 and AtMYB2 not only caused an ABA-hypersensitive phenotype but also improved osmotic stress tolerance.40) AtMYC2 is also a major transcription factor in JA-induced transcription, which suggests crosstalk between ABA and JA responses via the AtMYC2 transcription factor.41)
The EARLY RESPONSIVE TO DEHYDRATION 1 (ERD1) gene encoding a Clp protease regulatory subunit responds to dehydration and high salinity before the accumulation of ABA. This pattern of induction suggests that an ABA-independent pathway exists in the dehydration stress response of Arabidopsis.42) Analysis of the ERD1 promoter revealed two novel cis-acting elements, a 14-bp rps1-like region (CACTAAATTGTCAC) and a CATGTG motif, which are necessary for the induction of ERD1. Three NAC transcription factors, including RD26, bind the c-acting element to upregulate ERD1.43),44) Moreover, ZF-HD proteins are necessary for the NAC-activated expression of ERD1.45) This regulatory system is different from the DREB/CBF system in the ABA-independent induction of stress genes.
The homeobox protein HB6 functions as a negative regulator of the ABA response.46) HB6 interacts with ABA-inducible promoters with the CAATTATTA motif and interacts with the ABI protein phosphatase, which suggests its role in ABA response and stomata closure. HB6 is thought to function in the ABA response in drought stress.
Recently, we reported other transcription factors involved in abiotic stress responses. For example, a drought-inducible TCP13 gene is involved in morphological changes in leaves during drought stress.47) The TCP13 knockout mutant showed ABA-insensitive root growth and reduced dehydration-inducible gene expression. TCP13 is thought to function in both auxin and ABA responses in drought stress. Three bZIP transcription factors, bZIP60, bZIP17, and bZIP28, function in the unfolded protein response or ER stress responses to abiotic stress. Overexpression of bZIP60 improved salinity stress resistance.48) Double knockout mutants of bZIP17 and bZIP28 showed a short-root phenotype.49) Suppressor mutation in TAF12b recovered root growth of the double knockout mutant.50)
Transcription factors that are involved in abiotic stress responses are summarized in Table 1.
In the heat stress responses of various organisms, heat shock proteins (HSPs) function as molecular chaperons to protect cells from heat stress. Heat shock transcription factors (HSFs) function in the heat stress-responsive induction of HSPs. In plants, HSFA1s function as master regulators to regulate downstream transcription factors, including DREB2A and other HSFs (Fig. 3).13) We found that DREB2A is rapidly induced by heat stress in addition to drought stress responses.51) Domain analysis of DREB2A revealed that a negative regulatory domain (NRD) located in the central region of DREB2A is important for the regulation of DREB2A transcriptional activity.52) Transgenic plants overexpressing constitutively active forms of DREB2A (DREB2A CA) that lack the NRD show the enhancement of both stress-inducible gene expression and dehydration stress resistance. The stability of the DREB2A protein is regulated by modification of the NRD. DREB2A-interacting protein 1 (DRIP1) and DRIP2 were isolated as E3 ubiquitin ligases that mediate the ubiquitination of DREB2A.53) Ubiquitinated DREB2A is degraded through the 26S proteasome pathway in control conditions.
The DREB2A protein is regulated by other degradation pathways in heat stress conditions. BTB/POZ and MATH domain proteins (BPMs) encode substrate adaptors of the CULLIN3-based E3 ligase and bind to the NRD of DREB2A.54) The inhibition of phosphorylation in NRD by CASEIN KINASE 1 (CK1) stabilizes and activates DREB2A in response to heat stress.55) DREB2A mediates drought and heat stress signaling with the different regulatory mechanisms of NRD in DREB2A. The heat stress responses were reviewed in detail by Ohama et al. (2017).13)
DREB1s/CBFs bind to the DRE cis-acting element and act as master regulators in several cold-inducible transcription pathways. There are three DREB1/CBF genes in Arabidopsis: DREB1B/CBF1, DREB1A/CBF3, and DREB1C/CBF2, which are arrayed in this order. They are all rapidly induced by cold stress. Arabidopsis mutant plants in which three DREB1 genes are disrupted show decreased freezing tolerance (Fig. 3).
Two different types of transcription factors are involved in cold-inducible DREB1 expression, namely, the Calmodulin-Binding Transcriptional Activator (CAMTA) and circadian clock-related MYB-like transcription factors such as CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY) and REVEILLEs (RVEs). Plants recognize cold stress as two different types of signals, namely, rapid or gradual temperature decreases.56),57) CAMTAs respond to a rapid temperature drop, strongly inducing the expression of DREB1B and DREB1C. In contrast, clock-related MYB-like transcription factors regulate the expression of DREB1A and DREB1C in response to both rapid and gradual temperature decreases.14),56),57)
The MYC-like transcription factor ICE1 was reported to induce the DREB1A/CBF3 promoter based on the analysis of an ice1-1 mutant. However, DREB1A/CBF3 repression in the ice1-1 mutant was not correlated with ICE1 mutation. ICE1 is identical to SCRM1 in stomatal development but is not involved in the cold stress response. Therefore, many reports on the ICE1-DREB1A/CBF3 regulatory model in the cold stress response need to be revalidated without preconceptions.58)–60) More detailed analysis of the regulatory systems of cold-responsive transcription are reviewed in Kidokoro et al. (2022).14)
Integrated analyses of transcriptome and metabolomics analyses in Arabidopsis revealed connections between genes and primary metabolites in dehydration conditions. Metabolite profiling by gas-chromatography-mass spectrometry and capillary electrophoresis-mass spectrometry analyses revealed that ABA accumulates during dehydration, regulating the accumulation of various amino acids and sugars such as glucose and fructose.61) In particular, the dehydration-inducible accumulation of branch-chain amino acids, saccharopine, proline, and agmatine is correlated with the dehydration-inducible expression of their key biosynthetic genes, such as branch-chain aminotransferase 2 (AtBCAT2), lysine ketoglutarate reductase/saccharopine dehydrogenase (AtLKR/SDH), Pyrroline-5-carboxylate synthase 1 (AtP5CS1), and arginine decarboxylase 2 (AtADC2), respectively. These genes are induced by endogenous ABA in drought stress conditions.
Metabolites that are involved in abiotic stress response and tolerance are summarized in Table 2.
| Metabolite | Functions | Abiotic stress | References |
|---|---|---|---|
| Plant hormones involved in stress responses | |||
| ABA | Abscisic acid | Dry | 5, 66, 104 |
| ABA-Glu | Abscisic acid glucosyl ester | Dry | 66, 104 |
| JA | Jasmonic acid | Dry, Wound | 66 |
| JA-Ile | Jasmonic acid isoleucine | Dry | 66 |
| SA | Salicylic acid | Dry, Wound | 66 |
| Cytokinin | Kinetin, Zeatin | Dry | 66 |
| Metabolites involved in abiotic stress tolerance | |||
| Pro | Proline | Dry, ABA | 7, 61 |
| RFO | Raffinose family oligosaccharides (galactinol, raffinose) |
Dry, Cold, Heat | 7, 61, 62 |
| BCAA | Branch-chain amino acid (valine, leucine, isoleucine) |
Dry, ABA | 7, 61 |
| Saccharopine | Dry, ABA | 7, 61 | |
| Agmatine | Dry, ABA | 7, 61 | |
| GABA | γ-amino-butyric acid | ABA (−) | 7, 61 |
| Alanine | ABA (−) | 7, 61 | |
| Succinate | ABA (−) | 7, 61 | |
Plant hormones and metabolites involved in abiotic stress responses and tolerance are listed in this table and discussed in the text.
Dry: drought stress; Cold: cold stress; Heat: heat stress; ABA: abscisic acid regulated; Wound: wound stress. (−): negative regulation.
In contrast, the biosynthesis pathway of raffinose family oligosaccharides (RFOs) is controlled by the ABA-independent pathway. Galactinol synthase (GolS) is a key enzyme involved in galactinol and raffinose biosynthesis.62) RFO protects cells from oxidative stress.63) The AtGolS1 and AtGolS2 genes encoding galactinol synthase are induced by dehydration in an ABA-independent pathway. Overexpression of AtGolS2 increased drought stress tolerance, which has been applied to the development of drought-tolerant crops in the field.64) The γ-amino-butylate (GABA) shunt pathway to produce succinate from glutamate is negatively controlled by ABA. GABA, alanine, and succinate are negatively regulated by ABA. GABA decarboxylase GAD4 is involved in this process and ABA repression.61)
Metabolomics analysis of transgenic Arabidopsis overexpressing AtDREB1A/CBF3 revealed an ABA-independent metabolic network with similarity to the cold-induced metabolomics profile.65) Overexpression of AtDREB1A/CBF3 significantly increased cold stress tolerance. However, overexpression of AtDREB2A in transgenic plants increased their tolerance to dehydration stress but only slightly increased freezing stress tolerance.51) The increased tolerance to freezing stress in transgenic AtDREB1A-overexpressing plants depends on the accumulation of low-temperature-regulated metabolites, especially raffinose and galactinol.
Profiles of plant hormones were analyzed using dehydrated Arabidopsis plants in moderate dehydration stress.66) ABA is a major plant hormone in the drought stress response. Jasmonic acid isoleucine (JA-Ile) increased slightly in wild-type plants and significantly increased in nced3 mutants in response to dehydration. Salicylic acid and cytokines are decreased during the late dehydration response. In the drought stress response, there seems to be crosstalk between ABA and JA-Ile.
Plant hormones involved in abiotic stress responses are summarized in Table 2.
The perception and signal transduction in ABA responses have been thoroughly studied.3),5) ABA-insensitive mutants abi1 and abi2 were isolated, and both ABI1 and ABI2 genes encode Protein Phosphatase 2C (PP2C), which suggests an important role of protein phosphorylation in ABA signaling. Then, the protein kinase SnRK2 was shown to be a major protein kinase family in ABA signaling.67)–70) SnRK2 family genes have 10 members and are classified into 3 subclasses. Among them, 3 genes of subclass III SnRK2 play important roles in ABA signal transduction because their triple knockout mutant showed severe ABA-insensitive phenotypes.71),72) Subclass III SnRK2s interact with PP2Cs and phosphorylate AREB/ABF transcription factors to induce ABA-inducible genes.37),69) These observations support the important roles of SnRK2 and PP2C in ABA signal transduction pathways.
To understand ABA action, the identification of receptors is necessary. After long-term efforts by many groups, PYR/PYL/RCAR-type or START-type soluble ABA receptors were identified using chemical genetic and biochemical approaches.73),74) Biochemical and genetic analyses revealed that these proteins directly bind PP2Cs in the presence of ABA and inhibit their activity in an ABA-dependent manner. This finding indicates that ABA regulates cellular processes by modulating PP2C activity. Biochemical analysis showed that clade-A PP2Cs interact with ABA-activated SnRK2s and inactivate them efficiently through dephosphorylation at specific amino acid residues.75) Because SnRK2s can phosphorylate and activate some ABA-dependent transcription factors (see above), one of the ABA signaling pathways is now established from perception to gene expression, namely, PYR/PYL/RCAR-related receptor → clade-A PP2C → ABA-activated SnRK2 → AREB/ABF bZIP transcription factors (Fig. 4).75)–78)
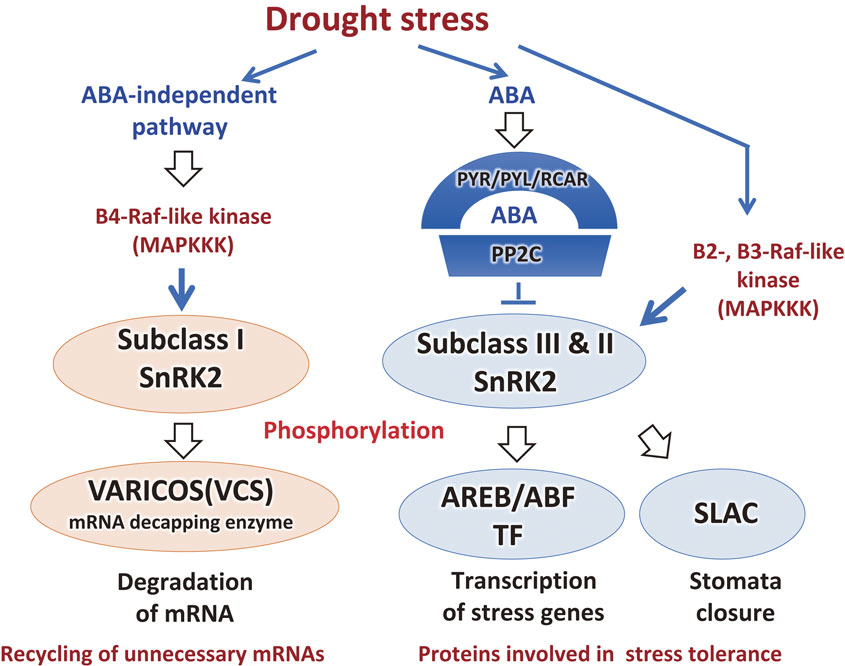
Cellular phosphorylation signaling in ABA-dependent and ABA-independent responses in drought stress. SnRK2 protein kinases are major protein kinases involved in drought stress responses. Subclass III SnRK2s function in the ABA-dependent pathway downstream of PYR/PYL/RCAR ABA receptors and the protein phosphatase PP2C. Upstream of SnRK2, Raf-like kinases function as activators of SnRK2 in drought stress conditions. Activated SnRK2s phosphorylate and activate AREB/ABF transcription factors to upregulate stress-inducible genes. In contrast, subclass I SnRK2 functions in the ABA-independent pathway to regulate downstream VCS, which regulates mRNA degradation. This process functions in the recycling of ribonucleotides for the induction of stress genes. Names and functions of genes for transcription factors (TFs) and signaling factors (SFs) are summarized in Table 1.
Phosphoproteomics analysis using triple mutants of subclass III SnRK2s revealed their downstream phosphorylated target proteins, which are bZIP transcription factors, including AREB1/ABF2, AREB3, and EEL.79) mitogen-activated protein kinases (MAPKs) MPK1 and MPK2 are targets of SnRK2. Unknown proteins, including SNS1, were identified as SnRK2 target proteins.
Subclass III SnRK2s are activated by MAPK kinase (MAPKKK) or B-type Raf-like kinase in Physcomitrellla patens.80) In Arabidopsis, B-type Raf-like protein kinases also function in the phosphorylation and activation of subclass III SnRK2. B2- and B3-Raf kinases are activated by osmotic stress in an ABA-independent manner, which indicates crosstalk between ABA-dependent and ABA-independent activation of SnRK2 (Fig. 4).81)–83) In contrast, subclass I SnRK2s are activated by dehydration stress in an ABA-independent pathway. Dehydration-activated subclass I SnRK2s regulate mRNA decay by phosphorylating VARICOSE (VCS), which is a scaffold protein of mRNA-decapping complexes.84) B4-Raf kinase phosphorylates subclass I SnRK2s. Therefore, the B4-Raf kinase – subclass I SnRK2 – VCS signaling system regulates the mRNA population to adapt to dehydration status. Recycling of unnecessary mRNAs in stress conditions is necessary for dehydration stress responses to synthesize mRNAs for stress proteins. Details of the phospho-signaling pathways in the drought stress response are provided by Soma et al. (2021).11)
Signaling factors and receptors in ABA response are summarized in Table 1.
Land plants sense water deficit status mainly in roots and monitor the reduction of water potential in the root vasculature. Then, the hydraulic stress signal in root cells is transmitted to leaves to mediate stomatal closure and stress-inducible gene expression to protect plants from dehydration status.15),85),86) The sensing mechanism of hydraulic stress in roots is complex and has not been clearly elucidated. Different types of sensing molecules have been identified as possible candidates for sensors of water deficit status, namely, osmosensors, mechanosensors, calcium channels, and ROS sensors (Fig. 5). Combinations of these sensors recognize water-deficit conditions to properly respond to drought stress. Molecular patterns of water-deficit stress are sensed by these different types of sensors, and the sensed patterns of water deficiency signals are then transmitted to cellular signaling networks to induce stress-specific gene expression profiles to achieve appropriate responses to dehydration status. There are different types of signaling pathways that properly control stress responses (Fig. 6).17)

Sensing system and cellular signal transduction in drought stress responses. Drought is a complex stress for plants and induces different types of stress responses, such as osmotic, mechanical, and oxidative stresses. These complex stress signals are sensed by different cellular sensors and transduced to downstream signaling networks, including phosphorylation, calcium, and ROS signals. Molecular patterns of stress signals activate certain genes, hormones, and transporters to induce proper stress responses to adapt to drought stress conditions. Names and functions of sensors, receptors and signaling factors are summarized in Table 1.

Interorgan signaling from roots to leaves in the drought stress response. Interorgan signals of water deficiency are transmitted via the vasculature from roots to leaves. Change in turgor pressure, production of ROS, and transient calcium signals function in the early drought stress response. Peptides and metabolites are produced in roots and transmitted to leaves via xylem. In leaves, ABA is mainly produced in the vasculature and guard cells in response to stress signals from roots and leaves. The CLE25 peptide is synthesized in roots in drought stress conditions, transmitted to leaves via xylem, and then induces ABA biosynthesis in leaves. Signal transport from leaves to roots has not been analyzed yet (shown as a dashed arrow). These were also discussed before.17) This figure is reprinted with modification from Ref. 17.
Histidine kinases have been analyzed as osmosensors in yeast and bacteria. In yeast, Sln1p encoding a histidine kinase functions as an osmosensor upstream of the HOG1 MAPK pathway.9) In Arabidopsis, histidine kinase 1 (ATHK1/AHK1) was shown to be a functional homolog of yeast Sln1p encoding an osmosensor histidine kinase.87) AHK1 was shown to function as a positive regulator in drought stress tolerance by the analysis of its mutant and overexpressors.88),89) Other AHKs, AHK2, AHK3, and AHK4 (CRE1), function as negative regulators through cytokinin regulation.90) There may be complex osmosensing systems, including AHK1 and downstream signaling MAPKs, in plants because plant MAPKs are activated not only by abiotic stress signals but also by biotic stress signals to control complex downstream events, including gene expression and physiological responses (Fig. 5).91)
Stretch-activated calcium channels are thought to function as osmosensor candidates in drought stress sensing. In plants, calcium-permeable mechanosensitive channels (MCAs) 1 and 2 sense osmotic changes in the plasma membrane to mediate Ca2+ influx to activate downstream cellular signaling.92),93) MCA1 is a homolog of yeast MID1 and functions as a Ca2+-permeable stretch-activated channel component in plants.94),95) MCA1 and 2 are other candidate osmosensors for complex drought stress responses. The calcium current across the plasma membrane is activated by various stress signals, including osmotic stress (Fig. 5). Ca2+ waves rapidly transmit stress information to distant tissues. REDUCED HYPEROSMOLALITY INDUCED Ca2+ INCREASE 1 (OSCA1) functions as a channel protein that mediates Ca2+ influx in the osmotic stress response.96)–98) The role of OSCA1 in stomatal closure is mediated only by osmotic stress-mediated Ca2+ influx in stomata. CALCIUM-PERMEABLE STRESS-GATED CATION CHANNEL 1 (CSC1)/OSCA1.2 functions as a hyperosmolality-gated calcium-permeable channel protein and mediates Ca2+ influx.99),100) Therefore, OSCA family membrane proteins can mediate Ca2+ influx in dehydration stress responses.
HYDROGEN-PEROXIDE-INDUCED Ca2+ INCREASE (HPCA) was reported as a sensor of H2O2 in guard cells.101) HPCA1, a leucine-rich repeat receptor-like kinase, is activated by H2O2 in the extracellular domain, and induces Ca2+ influx in guard cells for stomatal closure (Fig. 5). ROS signaling functions in response to various types of environmental stresses, including dehydration. ROS signaling is thought to function in complex drought conditions. The ROS wave is induced through respiratory burst oxidase D (RBOHD) and regulates stomatal closure of stressed leaves. ROS signaling has important roles in coordinating systemic responses in different leaves to extreme abiotic stresses.102),103) ROS are also involved in ABA signaling.
Sensors and receptors that are involved in abiotic stress perception are summarized in Table 1.
In salinity or drought stress conditions, accumulated ABA has pivotal roles in stress responses in plants through a drastic change in the gene expression profile and cellular processes. Our knowledge of the synthesis and metabolism of ABA has greatly increased.6),104) In particular, the identification of rate-limiting enzymes such as 9-cis-epoxycarotenoid dioxygenases (NCEDs) in the ABA biosynthetic pathway and P450 CYP707s in the ABA catabolic pathway has greatly enhanced our understanding of how plants control the level of this phytohormone. The genes for these two enzymes are activated by various stress treatments; NCED3 is strongly induced by dehydration and downregulated by rehydration, whereas CYP707A is further upregulated during rehydration, suggesting fine-tuning of the ABA level by the coordinated regulation of ABA production and breakdown systems.105),106) In addition, ABA is stored as an inactivated form, ABA glucosyl ester, in vacuoles and apoplasts. In dehydration conditions, ABA is released from the glucosyl ester form by β-glucosidase.107) Genes involved in ABA biosynthesis are strongly expressed in vascular tissues. Among them, the drought-inducible NCED3 gene is expressed strongly in vascular tissues and induces ABA accumulation in leaves.108) NCED3 is regulated by an NGA1 transcription factor with a B3 domain.109) Other genes involved in ABA biosynthesis are also expressed in vascular tissues, which means ABA synthesis mainly occurs in vascular tissues.6),17) In guard cells, genes involved in ABA biosynthesis are also expressed and function in stomatal closure in rapid response to water-deficit conditions (Fig. 6).
ABA is a mobile signal molecule in plant stress responses. Membrane transporters function in the intercellular transport of signaling molecules through cellular membranes. Different types of membrane transporters function as ABA transporters (Fig. 6).110) Among them, ATP binding cassette (ABC) transporter genes were shown to function as ABA transporters. There are 129 ABC transporter genes in Arabidopsis. Among them, a AtABCG25 Ds-tagged knockout mutant showed an ABA-sensitive phenotype.111) AtABCG25 encodes a half-size ABC transporter that belongs to the WBC subfamily. The AtABCG25 protein was localized on the plasma membrane. ABCG25 is significantly expressed in phloem companion cells. Biochemical analysis showed that AtABCG25 functions as an ABA exporter. Water loss of the ABCG25-overexpressing plants was much less than that of wild-type plants.112) AtABCG25-overexpressing plants showed higher water-use efficiency than wild-type plants.113) Another ABC transporter, AtABCG40, was shown to function as an ABA importer.114) AtABCG40 is expressed in guard cells. This is consistent with the hypothesis that ABA is a mobile signal between vascular tissues and epidermal tissues, including guard cells, that controls stomatal regulation. In addition, AtABCG22 was shown to function in stomatal closure.115) Its knockout mutant shows open stomata and a dehydration-sensitive phenotype. AtABC22 is expressed in guard cells. However, the molecule transported by AtABCG22 has not yet been identified.
In addition to the ABC transporter family, ABA transporters in other families of membrane transporters have been reported (Fig. 6). Arabidopsis NPF4.6 and several members of the NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER FAMILY (NPF) have been identified as ABA transporters.116)–118) NPF4.6 and NPF5.1 act as ABA importers. NPF4.6 functions in guard cells as well as vascular tissues, and NPF5.1 is expressed in leaf cells. These NPFs are involved in stomatal closure by regulating the amount of ABA. Arabidopsis thaliana DETOXIFICATION EFFLUX CARRIER 50 (AtDTX50), which belongs to the multidrug and toxin efflux transporter family, has been identified as another ABA transporter.119) The AtDTX50 protein has ABA exporter activity and localizes to the plasma membrane. AtDTX50 is preferentially expressed in both vascular tissues and guard cells. Atdtx50 mutant plants are more tolerant to drought with lower stomatal conductance. AtDTX50 can also modify ABA transport in both vascular and guard cells. Several ABA transport systems have been reported to date. Details of the different functions of these ABA transporter systems and their coordinated roles are discussed by Kuromori et al. (2018).110)
ABA related transporters are summarized in Table 1.
Precise analyses of the Arabidopsis genome have revealed the existence of many small open reading frames encoding peptides. These predicted small open reading frames are transcribed in plants, which suggests the existence of small peptides that function as hormones in various stages of plant development. Plant peptides mediate regulatory roles in cell-to-cell communication and long-distance signaling, such as phytohormones.7),16),120) As a functional feature of those mobile molecules, peptides are perceived by each peptide-specific receptor in each tissue, enabling peptide–receptor modules to precisely transmit environmental stimuli from sensing tissues to target tissues. The family of CLAVATA3/EMBRYO-SURROUNDING REGION-RELATED (CLE) peptides include well-characterized peptides active in developmental processes. We showed that one of the CLE peptides, CLE25, functions as a mobile root-to-shoot signaling molecule that modulates stomatal closure and the expression of stress-related genes in leaves in dehydrated soil (Fig. 6).121) CLE25 gene expression is enhanced in root vascular tissues in response to dehydration stress. Root vasculature-derived CLE25 peptide moves from the roots to the leaves and induces NCED3 expression to enhance ABA accumulation in the leaves. In leaves, BAM1 and BAM3 receptor-like kinases function as receptors of the CLE25 peptide and transmit the signal to induce NCED3 to accumulate ABA, which then regulates the stomatal aperture (Fig. 6). Taken together, plants sense water deficit conditions in their roots and integrate the information among distant organs to optimize stress adaptations and resistance in whole plant tissues.
Systematic analyses revealed that almost all CLE peptide genes are expressed in roots. Exogenous applications of synthetic CLE peptides inhibit root growth elongation, indicating that several CLE peptides, including CLE25, redundantly mediate the regulation of root development. Cle25 mutants show a normal root growth phenotype. The root-expressed CLE25 transmits information on water-deficit stress from the roots to leaves, and CLE25 functions in the adaptation to water deficiency to control plant growth in drought stress conditions. Precise reviews on mobile peptides were reported by Takahashi et al. (2020)7) and Yoshida et al. (2021).16)
To understand the complex mechanisms underlying plant responses to environmental stresses at the whole plant level, it is important to measure plant growth in environmental stress conditions by controlling growth conditions (Fig. 7). A quantitative phenotyping system facilitates automated measurements of plant growth by regulating the timing and strength of adverse environmental stimuli and conducting time-course observations of multiple parameters of plant growth in stress conditions. Several phenotyping systems have been established recently at different organizational scales. Automated phenotyping systems provide powerful tools for plant research by employing large-scale, high-throughput, and nondestructive systems to analyze time-dependent changes in plant growth (Fig. 7).122)–124) We combined the advantages of existing automated platforms to construct the RIKEN Integrated Plant Phenotyping System (RIPPS) with automated weighing and watering stations and continuous rotation of plants during the experiment to unify growth conditions.125) Two major benefits of this system are (1) the microenvironmental conditions experienced by individual plants are homogenized and (2) additional imaging devices can be easily set to any position. With the RIPPS, we can obtain digital images of individual plants in the daytime and during the nighttime dark conditions using infrared (IR) LED light (peak wavelength 950 nm). We used an IR camera to measure leaf temperature and a near IR camera to measure the water status of leaves. By using these different cameras, plant status can be measured more precisely in environmental stress conditions. Time-dependent changes in plant size and detailed analyses of temporal and spatial effects can be derived and quantified from time-lapse growth images. The RIPPS phenotyping platform is operated with the precise control of growth parameters, including temperature, light, nutrient availability, and water conditions. We used the RIPPS to establish quantitative evaluation methods for determining the growth responses and water use efficiencies of Arabidopsis plants in different water conditions.126) We also analyzed the involvement of ABA metabolism in water use efficiency using the RIPPS.125) These proof-of-concept studies demonstrate that the RIPPS accurately facilitates the measurement and evaluation of plant growth responses to precisely controlled hydric environments. By using a quantitative phenotyping system, we can analyze gene knockout mutants and natural genetic variants to perform high-throughput genetic mapping and large-scale analysis of gene expression pathways, metabolite levels, or plant hormone levels. Gene discovery of novel factors involved in abiotic stress responses can be achieved by improving phenotyping accuracy and throughput. A precise review on phenotyping of drought tolerance was provided by Kuromori et al. (2022).17)

Translation of basic knowledge on plant responses to drought stress for the application on molecular breeding of crops with higher yields in drought stress conditions. In cellular stress responses, many useful genes have been identified, such as transcription factors, protein kinases, ABA-related regulators, and many stress-inducible genes. Moreover, interorgan signaling molecules have also been discovered. By using these useful genes, genome editing technology and genetic modified organism technology can be used for the development of drought-tolerant crops. Phenotyping and data science will provide useful technology for systematic breeding and cultivation technologies for crops and vegetables in the future. The RIKEN Integrated Plant Phenotyping System (RIPPS) was reported by Fujita et al.125) and its operation can be seen in the website of RIKEN CSRS in the following URL. http://www.csrs.riken.jp/jp/virtualtour/yokohama/index.html?onstart=loadscene(scene9,null.MERGE,BLEND(1.2)).
The world’s food supply will need to increase due to the world’s population growth and economic growth, and food production will need to increase by more than 1.6-fold by 2050. Increasing the production of agricultural products has become an urgent issue. However, many fields have become dry and poorly nourished because of global warming. Therefore, there is a demand for the development of crops that are resistant to drought and have higher yields with less water (Fig. 7).2),12)
With such expectations, we have been conducting joint research with the Japan International Research Center for Agricultural Research (JIRCAS), the International Center for Tropical Agriculture (CIAT) in Colombia, the International Rice Research Institute (IRRI) in the Philippines, and the International Maize and Wheat Improvement Center (CIMMYT) in Mexico.127),128) This international collaboration was supported by a special fund (2007–2012) from the Ministry of Agriculture, Forestry and Fisheries (MAFF), Japan. We tested Arabidopsis and rice stress-related genes and various promoters. The stress genes used were transcription factors such as DREBs and AREBs, NCED3 in ABA synthesis, LEA proteins and metabolic enzymes involved in stress resistance. The promoters used are constitutive promoters and drought-inducible promoters. Among them, overexpression of the AtGolS2 gene in combination with the ubiquitin promoter rendered transgenic rice more resistant to drought and increased grain yield in drought conditions in the field. AtGolS2 is an Arabidopsis gene encoding galacitinol synthase, a key enzyme in RFO biosynthesis. RFO has a function in removing active oxygen.62),63) AtGolS2-overexpressing rice plants had higher relative water content in the leaves and drought tolerance. Cultivation of transgenic rice in a dry field in Columbia demonstrated that introduction of the AtGolS3 gene is effective in developing drought-resistant and high-yielding crops in dry fields.64),128) We also reported that other genes, such as transcription factors DREBs and AREBs, are also useful for the breeding of drought tolerance and higher yield in dry fields.128)
We collaborated with Embrapa (Brazilian Corporation of Agricultural Research) to test Arabidopsis stress genes in transgenic soybeans in greenhouses and dry fields. This project was supported by the Science and Technology Research Partnership for Sustainable Development (SATREPS) of the Japan Science and Technology Agency (JST) and Japan International Cooperation Agency (JICA) from 2009 to 2014.128) Transgenic soybean expressing Arabidopsis GolS2, DREB, AREB, NCED3, and other genes showed improved drought tolerance in the greenhouse. We also tested transgenic soybean lines in dry fields in Brazil.129) Some of these useful genes have also been applied to sugarcane, which has yielded good results and been applied to field trials.130)
Since 1989, we have been studying complex plant responses to abiotic stress conditions such as drought, heat, and cold stresses. We used functional genomics approaches to discover important genes and factors involved in gene expression and signal transduction in response to drought, heat, and cold stress conditions. We have identified key transcription factors in stress-inducible gene expression; important phosphorylation processes in stress signaling; key genes in ABA synthesis, degradation, and transporter; peptides involved in long-distance signaling; and metabolites for stress tolerance during more than 30 years of research and development. Some of the genes identified have been applied to improve the drought tolerance of crops and their productivity in drought conditions.
We have discovered sophisticated regulatory systems for plant responses and tolerance to drought and temperature stresses and their crosstalk in regulatory networks. It is still unclear how plants recognize complex environmental changes and stress to respond and adapt to survive in severe stressful conditions. We realized that plants have various types of sensors and signaling systems to recognize the molecular patterns of these stress signals in response to environmental changes and stresses. Molecular patterns of stress signals are transferred to properly induce downstream molecular events. In the coming decades, it will be important to understand the molecular patterns of stress recognition, signaling networks, and stress responses in complex environmental conditions (Fig. 7).17)
To understand plant responses to environmental stress, it is important to monitor plant phenotyping quantitatively by using imaging technology in combination with information technology. These phenotype data in various environmental conditions will be useful to evaluate plant performance in various stress conditions. Moreover, monitoring plant growth in controlled environmental conditions can provide useful tools for quantitative phenotyping. In combination with omics analyses such as transcriptomics and metabolomics analyses, plant status in certain conditions can be precisely described (Fig. 7). We can access various mutant resources, including gene knockout lines and ecotypes in Arabidopsis, which provide useful and precise data on plant responses to abiotic stress. By applying these methods to various plants and crops, we can analyze their responses at the molecular level. By integrating genomics and data science, we can describe profiles of plant responses and crop performance in various combinations of environmental stresses in the future.131)
We would like to thank all the members of our laboratories and collaborators for their valuable contributions. We also thank RIKEN, The University of Tokyo, and Japan International Research Center for Agricultural Sciences (JIRCAS) for their support of our research. We also thank the Japan Society for the Promotion of Science (JSPS), Bio-oriented technology Research Advancement Institution (BRAIN), and Japan Science and Technology Agency (JST) for funding our research. We apologize for not referencing all manuscripts on related topics due to space limitations.
Edited by Yoshinori OHSUMI, M.J.A.
Correspondence should be addressed to: K. Shinozaki, RIKEN Center for Sustainable Resource Science, 3-1-1 Koyadai, Tsukuba, Ibaraki 305-0074, Japan (e-mail: kazuo.shinozaki@riken.jp).

Kazuo Shinozaki was born in Utsunomiya city, Tochigi prefecture, Japan in 1949. He graduated from Osaka University in 1972. After entering the Graduate Course of Nagoya University he was awarded his Ph.D., under the supervision of Professors Reiji and Tsuneko Okazaki, in 1979. His thesis was on discontinuous DNA replication in T7 bacteriophage. He became a Research Associate at the National Institute of Genetics, Mishima, Japan, in 1978. Then, he became an Assistant Professor in the Department of Biology, Nagoya University, in 1983, and was promoted to Associate Professor at the Center for Gene Research, Nagoya University, in 1986. He determined the whole genome sequence of the tobacco chloroplast with Professor Masahiro Sugiura in 1986. He worked as a Visiting Scientist with Professor Nam-Hai Chua, Rockefeller University, U.S.A. from 1987 to 1989. He was promoted to Chief Scientist of the Plant Molecular Biology Laboratory of Tsukuba Life Science Center of RIKEN in 1989. From 1999, he led the plant functional genomics project in the Genome Sciences Center (GSC) of RIKEN. He became Director of the Plant Science Center (PSC) of RIKEN in 2005, and then Director of the Center for Sustainable Resource Science (CSRS) of RIKEN in 2013. From 2020, he became Senior Advisor for RIKEN CSRS. He was also Group Director of the Gene Discovery Research Group of CSRS. He was President of the Japan Society of Plant Physiologists (JSPP) from 2010 to 2011. He was elected as the University Professor of Nagoya University in 2017.
Kazuo Shinozaki was awarded the Tsukuba Prize in 2002 and the JSPP Award in 2009. He received a Person of Merit for Contribution to Culture and Medal with Purple Ribbon for Contribution to Science in Japan in 2016. He was selected as a corresponding member of the American Society of Plant Biologists in 2015. He was selected as an international member of the National Academy of Sciences of the U.S.A. in 2020. He was awarded the International Prize for Biology in 2020 and was awarded the Khalifa International Award for Date Palm and Agricultural Innovation by the U.A.E. in 2021. From 2014 to 2021, he was selected as a Highly Cited Researcher.
Kazuo Shinozaki has been studying plant regulatory gene networks in abiotic environmental stresses, especially drought stress. He is known for his studies on the transcriptional regulation of drought stress-inducible plant genes, signal transduction in drought stress responses and tolerance, and regulatory networks in responses to the phytohormone abscisic acid. Kazuo Shinozaki and Kazuko Yamaguchi-Shinozaki have been collaborating on these research topics since 1989. Kazuo Shinozaki also contributed to the Arabidopsis functional genomics project based on the collection of full-length cDNAs and tagged mutant lines for reverse genetics. He is working on phenotyping of plant growth in various environmental stress conditions.

Kazuko Yamaguchi-Shinozaki was born in Maebashi city, Gunma prefecture, Japan in 1954. She graduated from Japan Women’s University in 1977. She entered the graduate course of Tokyo Institute for Technology and was awarded a Ph.D. under the supervision of Professor Tsujiaki Hata in 1982. Her thesis was on the translational regulation of eucaryotic mRNAs. Then, she worked as a postdoctoral fellow at the National Institute of Genetics under the supervision of Professor Kin-ichiro Miura from 1982. She moved to the Center for Gene Research, Nagoya University, as a special researcher in 1983. Then, she worked as a postdoctoral fellow in Professor Nam-Hai Chua’s laboratory from 1987. She worked in RIKEN as a Special Postdoctoral Researcher from 1989. She became a Chief Researcher at the Japan International Research Center for Agricultural Research (JIRCAS) in 1993. She was promoted to Professor of the Department of Agriculture and Life Science, The University of Tokyo in 2004. From 2020, she has been an Emeritus Professor of the University of Tokyo. She has been Professor of Tokyo University of Agriculture since 2020.
Kazuko Yamaguchi-Shinozaki received the Tokyo Techno Forum Gold Medal in 2000. Then she was awarded the Tsukuba Prize and the MEXT Minister Award in 2002. She received with the JSPP Award from the Japanese Society of Plant Physiologists (JSPP) in 2009. She was awarded the MIDORI Academic Prize in 2018 by the Prime Minister of Japan. From 2014 to 2021, she was selected as a Highly Cited Researcher.
Kazuko Yamaguchi-Shinozaki is a plant molecular biologist mainly working on transcriptional regulatory networks in environmental stress responses, such as drought, cold, and heat stresses. She discovered key transcription factors in abiotic stress responses, including abscisic acid responses, and analyzed their upstream regulatory signaling factors in Arabidopsis. She has applied Arabidopsis and rice genes involved in stress tolerance for molecular breeding of crops, such as rice, soybean, wheat, sugarcane, and so on based on transgenic technology through international collaborations. Kazuko Yamaguchi-Shinozaki has been collaborating with Kazuo Shinozaki on these research topics on plant responses to abiotic stress since 1989.