2023 年 39 巻 3 号 p. 77-93
2023 年 39 巻 3 号 p. 77-93
The genome is carried by the complete linear sequence of DNA nucleotides packaged into all the chromosomes. During interphase, the chromosomes are extended and much of their chromatin exist as long threads while also maintaining specific three-dimensional architectures in the nuclear space. These interphase chromosomes are organized into multiscale three-dimensional structures, including chromosome territories, A/B compartments, topologically associating domains, and chromatin loops, that extend over a wide range of genomic distances, providing connections, for instance, between enhancers and promoters. This hierarchically organized genomic architecture is crucial for the regulation of gene transcription, which in turn is essential for the development and maintenance of various biological processes. This article reviews various aspects of spatial genome organization and their functions in gene expression and neural development. Furthermore, dysregulation of spatial genome organization in disease states, and the growing interest in new technologies to manipulate chromatin architecture are also discussed.
The sequencing of the human genome in 2001 [1, 2] provided an overview of the genome at the linear sequence level. The human genome is encoded by approximately 3.2 billion nucleotides of DNA, and most human cells, which are diploid, contain about 6 billion base pairs, divided into 46 chromosomes. These 6 billion base pairs, equivalent to about 2 m of linear DNA, are folded into three-dimensional (3D) structures and packaged into the nuclei, which are about 5–10 μm in diameter. Histone proteins compact DNA to form nucleosomes, which allows this packaging of the DNA into the microscopic nuclear space [3, 4]. Recent technical advances such as chromosome conformation capture (3C), which detect the interactions between genomic loci that are close to in 3D space of nucleus, led to new insights into the spatial organization of chromatin. Interphase chromosomes are organized at spatially hierarchical levels, from chromatin loops that allow associations between promoters and other regulatory elements such as enhancers over short- and long-range linear genomic distances, to chromatin domains, topologically associating domains (TADs), and A/B compartments; moreover, entire chromosomes themselves occupy defined regions of the nucleus, termed chromosome territories (Fig. 1). These hierarchical structures are essential for normal gene control, and disturbances in these structures have been implicated as factors contributing to gene dysregulation in disease. Here, we review the recent studies on 3D genome organization during neural development and on the disorganization of spatial chromosome architectures in disease states. Finally, we discuss how these new concepts stimulate our understanding to help address underlying disease mechanisms and lead to breakthroughs in the development of novel therapeutic targets.
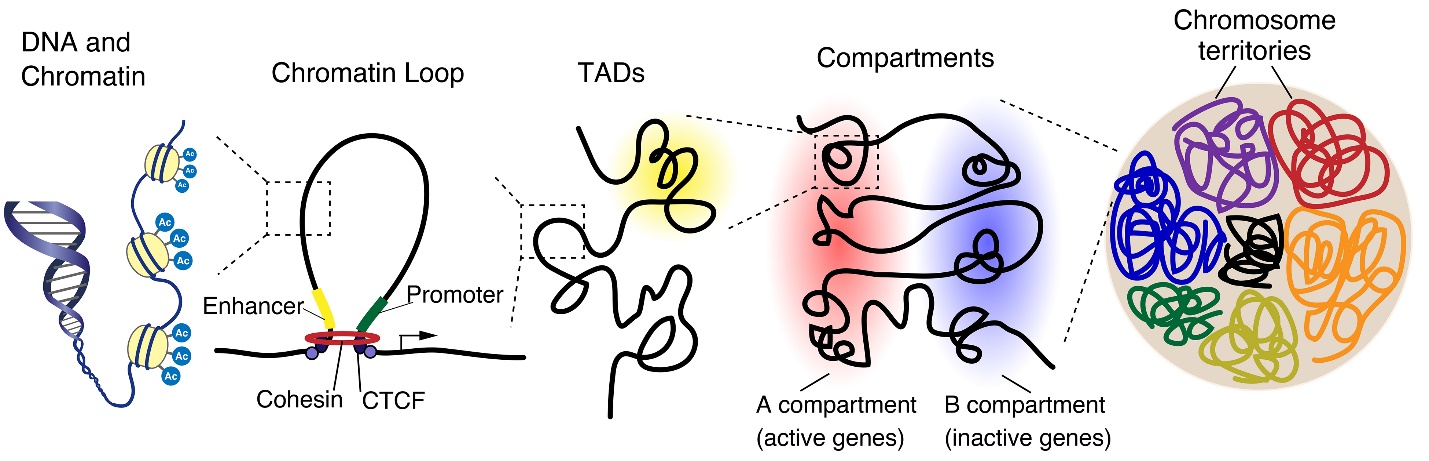
The genome is organized in a hierarchical manner, starting at a nuclear level containing all chromosomes down to individual chromatin fibers. Chromosomes occupy distinct regions in the nucleus, called chromosome territories, and generally avoid overlap. Each chromosome is separated into A and B compartments that include the transcriptionally active or inactive genes, respectively. Both A and B compartments involve topologically associated domains (TADs), whereby the genomic associations strongly occur within the domain. TADs are generally bordered by CCCTC-binding factor (CTCF), which connects to linearly distant DNA sequences and puts them into close three-dimensional proximity, leading to the formation of a chromatin loop.
Chromosomes are organized into hierarchical structures that play important roles in transcriptional regulation. This hierarchy of chromosomal structure is similar to aspects of protein structure, which starts with the folding of the amino acid sequence (primary structure) into secondary structures, such as alpha-helices and beta-sheets, and culminates in their organization into functional protein conformations [5].
In this section, we first provide a brief summary of the multiple layers of chromosome structural organization (Fig. 1), including epigenetic modifications to the linear genome, chromatin loops, TADs, A/B compartments, and 3D genomic locations in the nucleus [6–45].
During interphase, chromosomes tend not to intermingle but instead occupy distinct regions within the eukaryotic nuclear space. Microscopy-based approaches revealed that these chromosome territories are maintained through cell division, although the positions of chromosome territories can be flexible [46].
3C and derivative technologies, such as circularized chromosome conformation capture, chromosome conformation capture carbon copy (5C), and Hi-C, have helped reveal spatial chromatin architecture at a higher, mega-base scale resolution. Based on these techniques, chromatin organization can be classified into two major classes, A/B compartments and TADs.
Hi-C, which allows the complete detection of “all versus all” long-distance chromatin interactions across the entire genome, confirmed the presence of chromosome territories and also revealed intra-chromosomal compartmentalization into regions of open and closed chromatin, termed “A” and “B” compartments, respectively [47]. “A” compartments include genomic loci that are generally gene rich, transcriptionally active, and DNase I hypersensitive; conversely, loci found in “B” compartments are relatively gene poor, transcriptionally silent, and harbor heterochromatic sequences. The spatial segregation of A/B compartments was confirmed by microscopy-based methods [48].
Another major type of chromatin organization involves megabase-sized folding entities termed TADs. These were initially identified by Hi-C and 5C [49–51] and show a high frequency of interactions with regions outside the TAD boundaries. These domains were further characterized using Hi-C maps at 40-kb resolution [22] and are markedly smaller than A/B compartments—the median sequence size is 800 kb in TADs and 3 Mb in compartments. It has been proposed that restricting the interactions between genes and their regulatory sequences is one of the major functions of TADs [52, 53].
TADs contain sub-Mb scale genome organizations, such as sub-TADs and chromosomal loops [12, 54–56]. Generally, the loops between enhancers and promoters involve local interactions and are different from the long-range chromatin loops that are mediated by CCCTC-binding factor (CTCF), which originally identified as a transcription factor, and can also act as insulator. CTCF-mediated loops can facilitate enhancer–promoter interactions either by bringing enhancers and promoters together or by separating regions of active and silent chromatin [57, 58], which allows restricting promoter-enhancer interactions within TADs.
Studies that used chromatin contact mapping technologies have provided high-resolution views of DNA contacts associated with chromosome-structuring proteins [12, 59–65]. In mammals, TAD boundaries are usually demarcated by zinc-finger binding proteins, CTCF, and the cohesin complex [51, 66, 67]. The cohesin complex forms a ring-like structure that is comprised of four core subunits: the evolutionarily conserved structural maintenance of chromosomes (SMC) protein heterodimer, made up of SMC1α or SMC1β and SMC3; the double-strand-break repair protein, RAD21; and a stromal antigen homologue, SA1 or SA2 [68–70] (Fig. 2a, b). Cohesin cooperates with CTCF to form a chromatin loop and functions with the general transcriptional co-activator, the Mediator complex [71–75]. Some studies proposed that CTCF and cohesin promote “loop extrusion”, which contributes to TAD formation [76–80]. In this model, cohesin is loaded on to chromatin by cohesin loaders, including nipped-B-like protein (NIPBL) and MAU2, slides along the chromatin, and extrudes it outwards until it reaches the chromatin boundaries that are often formed by CTCF. Real-time visualization has recently confirmed that cohesin and its loaders induce genomic interphase DNA into loops by extrusion [81, 82]. Direct interaction of the N-terminal segment of CTCF with cohesin contributes to loop stabilizing activity [83, 84]. The study using cryo-electron microscopy provided insights into the probable mechanism of DNA entrapment by cohesin. NIPBL and DNA promote the engagement of the ATPase head domains of cohesin and ATP binding; thereafter, the hinge domains of cohesin dock onto its STAG1 subunit, creating a central tunnel to entrap DNA [85].
The deletion of the gene encoding for the cohesin-loading factor NIPBL or auxin-induced degradation of RAD21, one of subunits of the cohesin complex, results in the loss of CTCF-bound loops and of TADs, although A/B compartment segregation remains preserved [86, 87]. Similarly, acute depletion of CTCF by auxin-induced degradation also eliminated CTCF-bound loops and TADs in a dose-dependent fashion, while compartments remained largely unaffected [79]. In contrast, deletion of wings apart- like protein homologue (WAPL), which releases cohesin from chromatin lead to the extension of chromatin loops and strongly increased interaction frequencies between nearby TADs [88]. These results suggest that CTCF and cohesin are crucial for looping and TAD organization, whereas compartmentalization of mammalian chromosomes is regulated independently of local insulation by TADs. Moreover, several studies show that the ablation of CTCF or cohesin results in the loss of TADs but only moderately affects gene expression and histone modification [86, 87], suggesting that although transcriptional changes did occur, the regulatory potential remains preserved following the disruption of TADs.
Finally, in most cell types, large clusters of heterochromatin are enriched at the nuclear periphery. Lamina-associated domains (LADs), which are genomic regions that are in close contact with the nuclear lamina, are also thought to help organize chromosomes inside the nucleus and have been associated with gene repression [89, 90].
It has become possible to analyze the 3D structures of entire mammalian genomes at the single-cell level [91, 92]. This revealed that the structures of individual TADs and loops vary substantially from cell to cell, whereas A/B compartments, LADs, and active enhancers and promoters are organized consistently on a genome-wide basis in every cell.
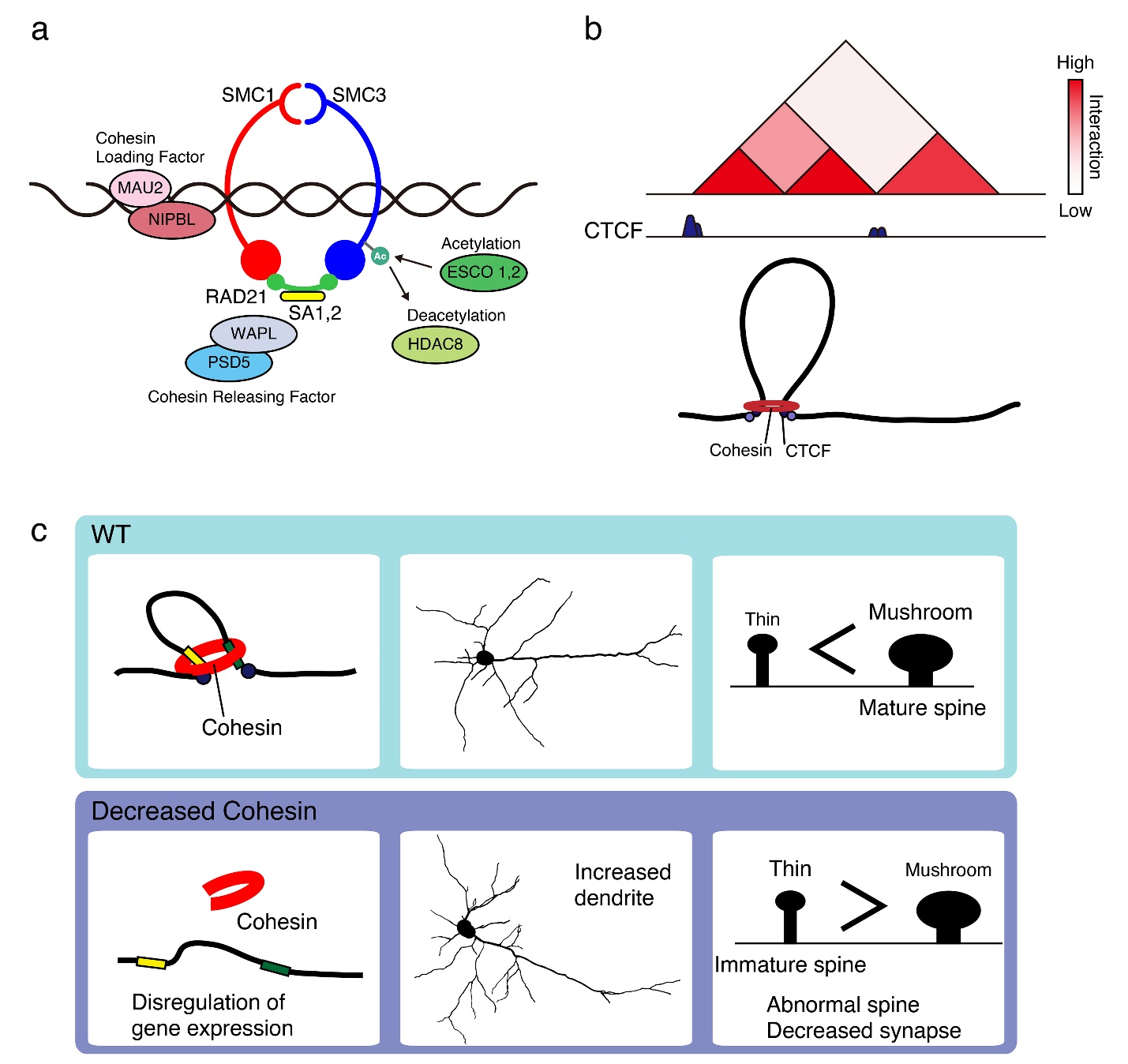
a) Cohesin and its associated proteins. The cohesin complex consists of four core subunits, SMC1, SMC3, RAD21, and SA1, 2. NIPBL and MAU2 complex loads cohesin on to chromatin, whereas WAPL and PDS5 release cohesin from chromatin.
b) Cohesin extrudes chromatin until it reaches TAD boundary by CTCF.
c) Heterozygous deletion of the cohesin subunit SMC3 impairs neuronal circuit formation and induces anxiety-like behaviors in mice.
Development is a complex process that involves changes in the expression patterns of various genes. Dynamic physiological changes in chromatin structure with epigenetic mechanism enable this stringently controlled spatio-temporal regulation of gene expression [93, 94]. Accumulating evidence has begun to clarify the roles of 3D genome organization in gametogenesis and early development [37, 95–104]. In this section, we focus on 3D genome dynamics and state-dependent changes in 3D genome organization during neuronal differentiation and development. With regard to neuronal differentiation, it seems that the “openness” of chromatin allows the cells to retain “stemness,” whereas chromatin structures get denser as they differentiate.
Global nuclear structure undergoes dynamic changes during sequential differentiation from embryonic stem cells (ESCs) to neural progenitor cells (NPCs) and then terminally differentiated neurons.
During the differentiation from NPCs to post-mitotic neurons, the number of chromocenters reduce, and they converge into larger clusters localized in the center of the nucleus [105–107]. These studies provide convincing evidence that an “open” chromatin structure is crucial for the pluripotency of these cells.
During cell differentiation, A/B compartments undergo dynamic switching as cells transition from ESCs to NPCs and then neurons. High resolution Hi-C analyses have revealed the progressive changes in differentiation stage-specific chromatin architecture both in mouse and human neurons. The interactions between active TADs become weak, whereas interactions in inactive TADs become stronger as mouse ESCs differentiate into NPCs and then neurons [108]. The compaction of nuclear chromatin domains seems to be a general feature of differentiating neurons and to contribute to the stable silencing of genes unnecessary for differentiated neurons.
Compared with A/B compartments, TADs appear to be relatively more stable during cell differentiation. TAD boundaries are stable during cell divisions and conserved among various cell types or lineages, although inter-TAD interactions and chromatin interactions within TADs can occur during cell differentiation [22, 33, 50, 51, 109–111].
Within TADs, chromatin structures seem to be more extensively reorganized locally thorough chromatin looping [112], for instance, those involved in promoter-enhancer interactions, which often occur in a developmental stage specific and cell-type specific manner [13].
Chromatin interaction analysis with paired-end tag sequencing has revealed the local chromosomal structures linked to the control of cell identity in ESCs [60]. During the course of neuronal differentiation, dynamic alterations of CTCF-mediated loops occur in both mouse and human developing brains [108, 113]. During the NPC to neuron transition, loops associated with cell proliferation, morphogenesis, and neurogenesis were lost, which is consistent with the commitment to a lineage change from the precursor stage towards a postmitotic neuronal identity. Similarly, during NPC to glia transition, loops associated with neuron-specific functions were lost, which is consistent with a non-neuronal lineage commitment. In addition, the loss of many shorter-range contacts and loops during the differentiation from NPCs to neurons was associated with concomitant increases in longer range (>100–200 kb) contacts in both humans and mice. These results provide insights into the relationship between transcriptional control of cell identity and local chromosome structure mediated by chromatin looping.
Gene expression is often associated with lamina-genome interactions—generally, genes that move away from the lamina are concomitantly activated, while genes that are located within LADs are transcriptionally inactive [89]. At least in four cell types, ESCs, NPCs, terminally differentiated astrocytes sequentially derived from ESCs, and mouse 3T3 embryonic fibroblasts, LADs are repressive chromatin features [114]. In contrast, other studies over the last decade have demonstrated that some chromatin at/in proximity to the lamina, especially those portions in proximity to nuclear pores, are euchromatic, with some highly expressed genes [115–117].
Thus, many studies have provided definitive evidence that links 3D genome organization and its function to gene expression, which can control developmental progression. However, the mechanisms underlying transcriptional regulation by chromatin architecture remain unclear.
Similar to the dynamic changes in chromatin structure during cellular differentiation, the role of 3D genome organization has been examined in the context of cell type and functions in retinal sensory neurons [118, 119]. Though heterochromatin normally resides at the nuclear periphery, whereas euchromatin situates toward the nuclear interior, it was found that this organization is inverted in rod photoreceptor neurons of nocturnal retinas [120–122]. The dense heterochromatin localized in the nuclear center may serve as collecting lenses to enhance light transduction efficiency in this case, which provides an example of how nuclear architecture is implicated in neuron function.
Spatial chromatin reorganization is also observed in the context of olfactory sensory neurons [123–129]. It has been reported that olfactory sensory neuron-specific and differentiation-dependent nuclear organization regulates the expression of the olfactory receptor [126]. During differentiation, olfactory sensory neurons select one out of the approximately 2,800 olfactory receptor alleles and execute the corresponding transcriptional program [130, 131]. The olfactory receptor genes converge into approximately five heterochromatin clusters located pericentrally in the nucleus of olfactory sensory neurons. This aggregation of olfactory receptor genes depends on the developmental decrease in levels of the lamin B receptor.
Collectively, these studies provide convincing evidence for the link between chromatin organization and neuronal function.
The central nervous system is a dynamic synaptic network that is highly influenced by the extrinsic environment. Extrinsic cues and the resulting synaptic activities drive transcriptional programs involving genes critical for proper neuronal maturation and neural plasticity. In this section, we focus on the activity dependent changes in 3D genome organization that regulate gene expression.
Early evidence reported by Billia et al. showed that the induction of long-term potentiation in rat hippocampal slice cultures causes the rearrangement of centromeric satellites [132]. Treating hippocampal neurons with N-methyl-D-aspartic acid (NMDA) increases centromere clustering, resulting in a decrease in the total number of centromere signals. Moreover, cultured rat hippocampal neurons change their nuclear structure in response to neuronal activity [133]. Wittmann et al. (2009) used 3D image reconstruction to find that many hippocampal neuron nuclei are highly infolded, with nuclear membranes that tuck inward and separate the nucleus into small and large compartments. The number of infolded nuclei are increased by NMDA receptor-induced calcium signaling, and nuclear calcium signals are stronger in smaller nuclear compartments than in the larger compartments of the same nucleus. Furthermore, activity-induced changes in the nuclear geometry are paralleled by increases in the phosphorylation of histone H3 on the serine 10 residue, suggesting a functional relationship between nuclear structure and transcriptional regulation. Three-dimensional tandem motion of chromocenters, called karyoplasmic streaming, is observed in the interphase nuclei of dorsal root ganglion neurons [134]. This motion occurs independently of concurrent motions in the cytoplasmic structure, and the speed of the motion is facilitated by neural stimulation with the nerve growth factor, gamma-aminobutyric acid, calcium ionophores, or calcium chelators [135], which represents activity-dependent dynamic changes in the global nuclear structure. Neuronal activation regulates synaptic structure and functions through the upregulation of some immediate early genes (IEGs), which are defined as genes that are rapidly and transiently induced (within 5 to 10 min of stimulation) by extracellular stimuli without the requirement for de novo protein synthesis [136–138]. Since IEGs often encode transcription factors, they can regulate a set of secondary response genes (SRGs), which are expressed in the order of hours in response to signaling and require new protein synthesis [139]. While there are probably several hundred IEGs, SRGs are far more numerous and are involved in various and cell-type specific functions in neurons. Given the rapid expression of IEGs without de novo protein synthesis, it has been extremely challenging to understand how they are transcriptionally regulated and whether specific 3D genome organization exists to control their expression, distinct from that of other genes that are expressed later and require de novo protein synthesis.
It has been reported that c-fos expression is tightly controlled by enhancer RNA (eRNA), which is non-coding RNA transcribed at active enhancer elements [140]; the authors showed that stimuli-induced promoter-enhancer loops mediate the transcriptional process. Further, the activity-induced Arc gene, which regulates synaptic plasticity, is also regulated by promoter-enhancer interactions. Immediate early genes, including Arc, are poised by RNA polymerase II at the downstream transcription start site. The loop-bound eRNA recruits the negative elongation factor complex, which inhibits transcription and liberates the target promoter, leading to the rapid induction of neuronal IEGs.
Recent studies have been revealed the links chromatin loop to activity dependent gene expression. Proximity ligation-assisted ChIP-seq (PLAC-seq) identified long-distance interactions between activity-dependent gene promoter with enhancer upon neural stimulation [141]. The core cohesin subunit Rad21 is required for activity dependent transcription, and the occupancy of Rad21 at enhancers and promoters correlated with changes in H3K27ac upon neural stimulation. Conditional CRISPR knockout of Rad21 in adult mouse granule neurons significantly reduced enhancer–promoter interactions undergoing delay motor learning. These results suggest the relation of activity dependent changes in chromatin loops and transcription to drive brain function.
A study used high-resolution 5C, which allows the detection of chromatin interactions between all selected fragments within a given region (typically on the megabase-scale), and Hi-C to reveal how activity-dependent enhancers are temporally connected via long-range chromatin loops to regulate gene expression during a wide range of neuronal activity paradigms. IEGs, including Fos and Arc, connect to activity-dependent enhancers via singular short-range loops that form within 20 min after neuronal stimulation, whereas the SRG Bdnf engages with both pre-existing and activity-inducible loops that form within 1–6 h [142]. Moreover, activity-dependent loops form prior to the peaking of mRNA levels of IEGs [142]. Neural activity has been known to regulate gene expression, but the causative mechanism is still unclear. These observations provide the possibility that activity-dependent nuclear remodeling contributes to activity-dependent changes in gene expression in neurons. Flexibility of the 3D genome structure might allow the rapid control of neuronal activity-induced gene expression, resulting in the corresponding cellular function.
In addition to evidence supporting the crucial role of spatial chromatin architecture in gene expression, findings from numerous studies have noted its association with mutations that occur outside protein coding region, suggesting the importance of regulatory elements such as promoters and enhancers in normal development [143]. In other words, dysregulation of chromatin architecture is frequently linked to diseases. Indeed, as mentioned above, deleterious mutations in the genes that encode chromatin architectural proteins such as CTCF and cohesin cause various developmental abnormalities.
Deleterious mutations in cohesin core subunit- or cohesin-related genes cause the multisystem developmental disorder Cornelia de Lange syndrome (CdLS) [144–146]. Mutations in the gene for cohesin loader, NIPBL were first identified in CdLS, followed by mutations in the genes for the cohesin subunits SMC1A, SMC3, and RAD21 [147–150]. Mutations in regulatory factors, HDAC8, which regulates SMC3 acetylation and cohesion cycling, BRD4, and ANKRD11 were also detected [151, 152]. Individuals with CdLS show diverse symptoms, including intellectual disabilities, anxiety, attention deficit hyperactivity disorder, and autism-like characteristics [153, 154]. Smc3 heterozygous deficient mice show increased dendritic complexity and decreased spine density in cortical neurons [155, 156]. Neuron-specific Smc3 deletion mice exhibit the same phenotype as the global heterozygous knockout mice, indicating that cohesin function in postmitotic neurons is required for proper neuronal network formation (Fig. 2c). These mice also demonstrate increased anxiety-related behaviors that are consistent with the symptoms of CdLS. A gene-ontology analysis following RNA sequencing revealed altered gene expression in the cortices of Smc3 heterozygous deficient mice compared to that in wild-type mice. Loss of cohesin function causes disruptions in chromatin loops, with subsequent effects on transcriptional regulation [66, 157–159]. Conditional CRISPR knockout of Rad21 in granule neurons disrupts enhancer-promoter interactions and the tactile startle response, which suggests that specific loops mediated by cohesin may be required for motor-learning [141]. These observations link 3D genome architecture to brain functions.
Studies on in vivo ablation of CTCF have also suggested important roles for chromatin architecture in brain functions. De novo mutations in CTCF cause intellectual disability [160], and neuron-specific CTCF knockout mice exhibit decreased dendritic arborization and spine density. These mice also show postnatal growth retardation and abnormal behaviors. Furthermore, ablation of CTCF disturbs gene transcription, including that of the protocadherin genes [161].
Kilobase-resolution of Hi-C analysis allows to detect specific chromatin loops between cis-regulatory elements [51, 55]. Hi-C mapping in induced pluripotent stem cell-derived NPCs revealed cell type-specific chromatin interactions in the schizophrenia (SZ) risk locus [113]. Compared with those in glia and other non-neuronal cells, larger numbers of cell-type-specific chromosomal contacts anchored in the risk locus were detected in neurons and NPCs. Since the authors also showed loss of short-range loops and the overall contraction of the spatial genome space during the NPC to neuron differentiation, chromatin interactions associated with SZ risk may increase as neurons differentiate. In addition, the SZ risk-related chromosomal connectome specific to NPCs or neurons showed coordinated gene expression and proteomic interactions. Thus, developmental chromatin interactions at the SZ risk-related locus, which occur more frequently in neurons than in other cell types, reflect the cell type–specific vulnerabilities in spatial genome organization.
However, Kilobase-resolution of Hi-C analysis is often challenging, since it requires sufficient number of cells (1–10 million) and high sequencing depth at billion-scale. A new low input (50–100k cells) “easy Hi-C” overcomes the limitations of Hi-C by using a biotin-free strategy to enrich ligation products [162]. Combined with a rigorous Hi-C bias-correction pipeline (HiCorr), that substantially improved the mapping of sub-TAD chromatin loops at fragment resolution, reveals cell-type specific chromatin loops or enhancer aggregates during neural differentiation (Fig. 3). Chromatin loops, but not compartments can be hallmarks of neural differentiation and neural functions. In addition, these new toolsets concluded that Hi-C loop outperforms expression quantitative trait locus (eQTL) in defining neurological genome-wide association study (GWAS) target genes. Some known brain GWAS loci lose pre-formed chromatin loops during neural differentiation. For instance, CTCF loop connecting the GWAS locus in the third intron of CACNA1C, which is strongly associated with SZ and bipolar disorder [163], to the CACNA1C promoter is specifically detected in human induced pluripotent stem cells (hiPSC). The loop weakens towards neurogenesis (i.e. in hNPC and hNeuron) when the gene is upregulated. Transcription elongation possibly cause this loop disruption [164]. These findings provide the working model that the GWAS locus gains a gene regulatory potential when it connects to gene promoter mediated by CTCF loop, and genetic variants in the risk locus may affect gene expression. Thus, high-resolution 3D genome analysis is efficient approach to elucidate the disease etiology.
Dysregulation of activity-dependent signaling contributes to the pathogenesis of neurodevelopmental and neuropsychiatric diseases [165]. Recently, risk-associated single-nucleotide variants (SNVs) associated with diseases, such as autism spectrum disorders (ASDs) and SZ, have been shown to colocalize with distinct classes of activity-dependent looped enhancers [142]; different features of schizophrenia-associated SNVs, which are connected to downregulated genes after synaptic activity, and ASD-associated SNVs, which connect activity-inducible enhancers to activity-upregulated target genes, were observed. These results suggest the possible existence of disease-specific alterations of activity-dependent looping, which lead to disease-specific neuronal phenotypes.

The distal GWAS locus related to schizophrenia and bipolar disorder is recruited to the CACNA1C promoter by CTCF-mediated loop only in hiPSCs when its expression level is low but detectable. The loop gets weaker in hNPCs and hNeurons when CACNA1C expression is upregulated.
Although we have focused on the 3D organization of chromatin structure during neural differentiation in this review, it is a crucial phenomenon important across the entire lifespan [166, 167]. Our understanding of the role and regulatory mechanisms of spatial chromatin structure and its effects on gene expression and biological and physiological functions has progressed significantly. However, as new findings emerge, they raise further questions regarding the mechanism and function of the 3D genome organization in gene expression and physiological functioning. One prominent question is discerning cause and consequence of the spatial alteration of chromatin architecture. As discussed above, ablation of CTCF or cohesin has limited effects on gene expression and histone modification even though it causes TAD loss [86, 87], suggesting that transcriptional regulation might be preserved even if TADs are almost completely disrupted. On the other hand, deletion of the genes encoding for CTCF or cohesin causes severe deficits in neural circuit formation and behavior. Taken together, it is intriguing to hypothesize that the disorganization of chromatin domain interactions, and not of transcriptional regulation, could be a causal effect of neurodevelopmental disorders. Furthermore, in a single cell ATAC-seq study in the developing human forebrain, cell type-specific changes in chromatin accessibility during corticogenesis were detected, and cell type distinctions beyond transcriptional definitions could be resolved [168]. Recent technical advance by developing Simultaneous High-throughput ATAC and RNA Expression with sequencing (SHARE-seq) enables to assess combination measures of chromatin accessibility and gene expression within the same single cell. SHARE-seq identified cis-regulatory chromatin interactions and showed chromatin accessibility at domains of regulatory chromatin precedes gene expression during lineage commitment [169]. These findings suggest the causal changes in chromatin accessibility may induce transcriptional changes. The causes of chromatin folding and its consequences for chromatin function in gene expression and biological functions should be discussed more in light of future studies and technical advances.
Aberrant 3D genome organization has been detected in neurological diseases such as SZ and ASD. In addition, mutations in the chromosomal architectural proteins, such as CTCF and cohesin impairs neural development and brain functions. Therefore, techniques for the manipulation of 3D genome organization, such as forced chromatin looping, could be novel therapeutic targets for neurodevelopmental diseases. Such technical improvements promise to help in unveiling the principles of 3D genome organization and its functions and in the development of novel approaches to repair the 3D genome disorganization in disease states.
Conflict of interest
The author declares there are no conflicts of interest.