2013 Volume 230 Issue 3 Pages 171-176
2013 Volume 230 Issue 3 Pages 171-176
Single Nucleotide Polymorphisms (SNPs) can genetically predispose individuals for certain diseases and therefore are of clinical significance. Myocardial infarction (MI) was investigated in large genetic association studies revealing novel SNPs associated with MI. rs4977574 is a non-protein coding SNP (A>G) that is located in proximity of cyclin-dependent kinase inhibitor 2A and B genes on chromosome 9p21.3. rs4977574 has been recently found to be associated with the early-onset of MI, and rs4977574 is characterized by a guanine nucleotide (G) instead of an adenine nucleotide (A). rs4977574 has been reported to increase the risk for MI by 28%. In this study, we developed a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method for detecting rs4977574 in Turkish population that consisted of 28 controls without previous MI record and 44 patients with MI. An intergenic genomic region containing the target SNP was amplified by PCR using patient’s genomic DNA. Amplified DNA fragments were digested with a restriction enzyme, HhaI that cuts the amplified sequence if only the sequence has GCGC that carries rs4977574. After digestion with HhaI, DNA fragments were visualized in order to detect genotypes. PCR-RFLP revealed that the frequency of rs4977574, the MI-associated allele (G), was 56.8% (25/44) in patients with MI and 33.9% (9.5/28) in controls; the frequency of rs4977574 in patients with MI was significantly higher compared to controls (P = 0.027). Importantly, for the first time in this study, we have developed a novel PCR-RFLP method to detect the presence of rs4977574.
Occurrence of myocardial infarction (MI), commonly known as heart attack, may be triggered by many factors including physical or psychological stress (Wilson et al. 1998). In addition to these factors, genetic factors are involved in the occurrence of myocardial infarction and MI is currently one of the most common health problems as a cause of mortality worldwide (Nora et al. 1980; Rosamond et al. 2008). Myocardial infarction is generally observed in people older than 65 years old, on the other hand, a small proportion of patients corresponding to 5 to 10% of all patients have this disease at younger ages (Rissanen 1979; Rosamond et al. 2008).
Genomes differ by small variations of single nucleotide polymorphisms (SNPs) that are differences of a single nucleotide among individuals. Genomic variations are examined in a large group of patient and control individuals in genome-wide association studies (GWAS) in order to find out SNPs or larger genetic events involved in disease occurrence (Pearson and Manolio 2008; Manolio 2010). Until today, hundreds of SNPs were identified to be associated with many major diseases (Johnson and O’Donnell 2009).
Heritable factors may be responsible for at least 30% of the risk for coronary artery diseases (CAD) (Marenberg et al. 1994). CAD and MI were investigated in large genetic association studies and GWAS revealed novel SNPs for these diseases especially on chromosome 9p21.3 (Helgadottir et al. 2007; McPherson et al. 2007; Samani et al. 2007). rs4977574 is an SNP on chromosome 9, band p21.3. rs4977574 is characterized by a guanine (G) nucleotide as the risk allele and the alternative allele is characterized by an adenine (A) nucleotide. It is a non-coding SNP that is located in a gene-free region, and the nearby genes encode cyclin-dependent kinase inhibitor 2A (CDKN2A) and cyclin-dependent kinase inhibitor 2B (CDKN2B). rs4977574 has been recently found to be associated with the early-onset of MI and affect the expression level of a nearby CDKN2B gene (Schunkert et al. 2008; Kathiresan et al. 2009). The frequency of rs4977574, MI-associated allele (G), is about 46-56% for Caucasians (Schunkert et al. 2008; Kathiresan et al. 2009; Schunkert et al. 2011). The method for the identification of this SNP is mostly high throughput SNP microarrays. The involvement of rs4977574 in CAD was further validated by a large-scale association study with a p value of 1.35 × 10−22 (Schunkert et al. 2011).
Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) is a method used for genotyping of sequence differences and genomic variations such as SNPs between subspecies. In PCR-RFLP, a marker region is amplified by polymerase chain reaction (PCR) and the amplified DNA fragment is digested by specific restriction enzymes. Presence or absence of certain DNA fragments after the restriction digestion reveals the genotype for the target SNP. There are numerous studies utilizing PCR-RFLP in order to investigate the association between MI and gene polymorphisms in genes such as arginase I, apolipoprotein CIII, SDF1 and endothelial nitric oxide synthase (Jo et al. 2006; Luan et al. 2010; Sediri et al. 2010, 2011).
In this study, i) for the first time, we developed a novel PCR-RFLP method to identify the occurrence of the SNP, rs4977574, ii) applied the method in a group of patients and controls for the first time in Turkish population and iii) showed increased frequency of rs4977574 in patients with MI.
The study population was composed of 44 patients with ST elevation MI, recruited from the Erciyes University Faculty of Medicine, Department of Cardiology, and 28 Turkish age-matched control subjects without documented coronary artery disease. The study protocol was approved by the Erciyes University Ethical Committee. Written informed consent was obtained from all subjects.
DNA isolationDNA isolation from blood samples of patients were performed by using GF-1 Blood DNA Extraction Kit (Vivantis, Malaysia) according to the procedure of the kit. Purified DNA samples were kept frozen at −20oC in nuclease-free distilled water.
Primer design and PCRAn intergenic genomic region of 301 bp on chromosome 9p21.3 was amplified by PCR using the genomic DNA isolated from blood samples of patients. NCBI reference number for the source contig sequence is NT_008413.18. Amplified genomic region contains a single nucleotide polymorphism, rs4977574. Novel primers were designed as the following: rs4977574-Forward: ATAGGGGTTATGGGAAATGC and rs4977574-Reverse: AAACCTAAAAGGGCTTGCTGA (Fig. 1).
In PCR reactions; 25 µl 2 × master mix (Vivantis, Malaysia), 4 µl 20 pmoles rs4977574-Forward and Reverse primers, 2 µl genomic DNA and 19 µl distilled water were used for a 50 µl reaction mix. PCR conditions were as following: i) hot start at 95°C for 5 minutes, ii) 35 cycles of denaturation at 94°C for 1 minute, annealing at 51°C for 1 minute and extension at 72°C for 1 minute and iii) final extension at 72°C for 10 minutes. Amplicons obtained by PCR amplification were run in 2% agarose gel at 120 V for 30 minutes and visualized in Gel Logic 212 Pro Jel (Carestream, USA). 100 bp DNA marker was used (Solis BioDyne, Estonia). Amplicons in PCR reactions were directly used in the restriction digestion analysis.

The Nucleotide Sequence of the Amplified Genomic Region.
Italic bold sequences with arrows at the beginning and the end of the sequence are forward and reverse primers, respectively. The nucleotide sequence marked with the square frame is the restriction site for the enzyme, HhaI. The nucleotide G with bigger font in the square frame is the SNP, rs4977574.
PCR products were digested with the restriction enzyme, HhaI (Vivantis, Malaysia). Restriction reactions were performed in 12 µl using about 0.5-1 µg DNA with following conditions: 10 units of HhaI at 37°C for 2 hours in 1 × Buffer 4 and 1 mg/ml BSA provided in the enzyme kit. Digestion products were separated in 2% (w/v) agarose gels, visualized in Gel Logic 212 Pro Jel and photographed.
Statistical AnalysisAll statistical analysis was performed using the SPSS 15.0 version (SPSS Inc. Chicago, IL, USA). Frequency of rs4977574 allele in patients with MI and controls was calculated. Chi-square test was used to calculate two-sided p value to decide whether there is a statistically significant difference between patients with MI and control groups in terms of the frequency of rs4977574 allele. P < 0.05 was considered statistically significant.
The characteristics of the patients and control subjects are shown in Table 1. The mean age was 60.1 ± 10.4 years (range 40-84 years) in patients and 58.4 ± 8.4 (range 36-75 years) in control subjects. The proportion of smokers was higher in patients than the control group (p < 0.001). On the other hand, there were no significant differences between groups for diabetes, hyperlipidemia, hypertension and the family history of cardiovascular disease (p > 0.05).
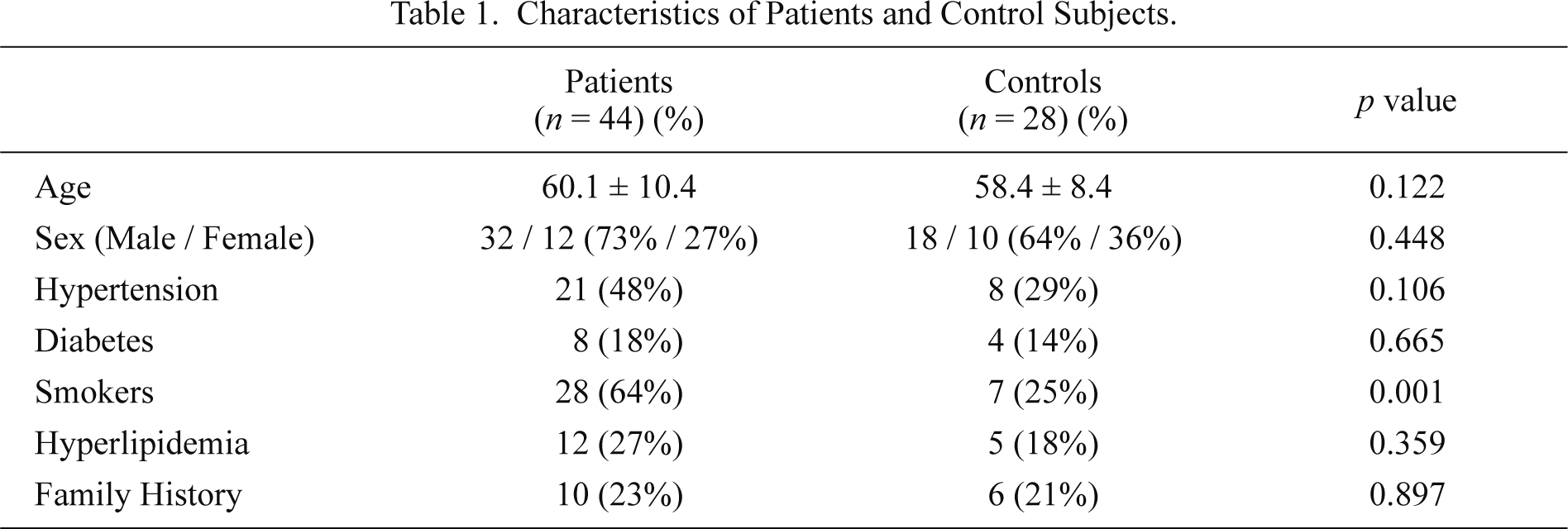
Characteristics of Patients and Control Subjects.
Primers were designed to amplify a 301 bp genomic region on chromosome 9p21.3 (NCBI Reference Sequence: NT_008413.18). This region includes the SNP, rs4977574. Primers, the position of the SNP and amplified sequence were depicted (Fig. 1). Genomic DNA was isolated from blood samples of 44 patients with MI and 28 controls. Genomic DNA for each patient was used as a template to amplify a PCR product with a size of 301 bp by using designed primers. Amplified DNA fragments were visualized on an agarose gel (Fig. 2). After the confirmation of the successful amplification of the target sequence, PCR reactions were directly used in restriction enzyme digestion. Amplified DNA was digested with the restriction enzyme, HhaI. PCR amplified fragments that were digested with HhaI, were run and visualized on an agarose gel. HhaI recognizes the sequence GCGC and cuts the amplified sequence if only the sequence has the SNP risk allele. The digestion does not occur if the SNP is not present and sequence is ACGC. As a result of digestion, DNA fragments of 187 bp and 114 bp were observed as two separate DNA bands in samples positive for the SNP risk allele (G) of rs4977574 (Fig. 3). Restriction digestion profile for each sample was analyzed. An example of PCR-RFLP with nine samples was shown. A complete digestion was observed for two homozygous samples (samples 3 and 6) and a partial digestion was obtained for five heterozygous samples (samples 1, 4, 7, 8 and 9). No digestion was observed in two samples negative for rs4977574 (samples 2 and 5) (Fig. 3).

PCR Amplification Results.
The intergenic genomic region containing the target SNP was amplified by PCR and a 301 bp PCR product was obtained. Amplified DNA fragments were run in 2% agarose gel and visualized. Presence of PCR products was confirmed. Each number represents a patient sample. Arrows represent the band sizes of the 100 bp DNA marker. N = Negative Control, M = DNA Marker, bp = base pair.

PCR-RFLP for rs4977574.
PCR products of 301 bp obtained from each of 9 samples were digested with HhaI (sample numbers with the asterisk). A negative control for each sample that contains the PCR product but does not contain the restriction enzyme was included (sample numbers without the asterisk). Restriction fragments of 187 bp and 114 bp were obtained for the samples that are positive for the SNP, rs4977574 and DNA fragments were observed as two separate DNA bands between 100 bp and 200 bp bands of the marker. A complete digestion was observed for homozygous samples (samples 3 and 6) and a partial digestion was obtained for heterozygous samples (samples 1, 4, 7, 8 and 9). No digestion was observed in samples negative for rs4977574 (samples 2 and 5). Arrows represent the band sizes of the 100 bp DNA marker. M = DNA Marker and bp = base pair.
Overall 44 samples of patients with MI and 28 samples of controls were analyzed. Among 44 patients with MI, 14 were homozygous, 22 were heterozygous and 8 were negative for rs4977574. Among 28 controls, 4 were homozygous, 11 were heterozygous and 13 were negative for rs4977574 (Table 2). The frequency of SNP risk allele (G), rs4977574, in patients with MI was 56.8% and frequency of rs4977574 in controls was 33.9%. The frequency of rs4977574 in patients with MI and controls was significantly different (P = 0.027) (Table 2).

Distribution of alleles carrying rs4977574 in patients with MI and Controls.
Genetic variations affecting MI have been studied extensively and especially genome wide association studies (GWAS) revealed numerous single nucleotide polymorphisms associated with MI. High throughput SNP arrays for the analysis of approximately a million of genome-wide SNPs, were utilized in GWAS. On the other hand, in contrast to expensive methodologies, small scale studies in local study groups have been performed using more conventional and economical methods such as PCR-RFLP (Fan et al. 2006). In Turkey, there are a few studies utilizing PCR-RFLP to analyze the role of gene polymorphisms in MI (Taşkiran et al. 2009; Yilmaz et al. 2009; Ergen et al. 2011). Another study from Japan investigated several polymorphisms in five different ethnic groups including Turks (Fujihara et al. 2006). Frequency of SNPs and significance of SNPs in MI need to be studied more extensively in Turkey.
rs4977574 is one of the SNPs that was found to be associated with the early-onset of MI in GWA studies that were performed with a large number of patients and controls. GWA studies with this SNP were performed with Caucasian and Hispanic populations, but frequency of rs4977574 and the effect of rs4977574 on MI have not been studied in Turkish population (Kathiresan et al. 2009; Qi et al. 2011; Schunkert et al. 2011). To the best of our knowledge, there is not a published PCR-RFLP method to study the presence of rs4977574. In order to study the frequency of rs4977574 in Turkish population and to investigate its role in patients with MI, a novel PCR-RFLP assay was developed in this study. In this assay, genomic DNA from patients and controls was used to amplify a genomic region that contains target SNP allele. Amplified DNA fragment was digested with the restriction enzyme, HhaI to detect the presence of SNP. In patients carrying the SNP risk allele (G), rs4977574, restriction digestion resulted in two DNA fragments and in patients not carrying the SNP risk allele (A), restriction digestion did not result any fragment. Additionally, PCR-RFLP assay was able to discriminate between patients who were homozygous or heterozygous for the SNP risk allele. A complete digestion occurred if the patient was homozygous, and an incomplete digestion occurred when the patient was heterozygous for the risk allele. In conclusion, the newly developed PCR-RFLP provides a fast, practical and economic tool for the study of rs4977574 in clinical and research laboratories.
Interestingly, the frequency of rs4977574 in our patients with MI is 56.8% and the frequency of rs4977574 in our control group is 33.9%. The frequency of rs4977574 is higher in patients with MI compared to controls without any previous MI record (P = 0.027). This is in accordance with published results for Caucasian and Hispanic populations (Kathiresan et al. 2009; Qi et al. 2011; Schunkert et al. 2011). In a study published by Myocardial Infarction Genetics Consortium in 2009, the frequency of rs4977574 was found to be 56% and with each copy of the rs4977574 allele, risk for MI was reported to be increased by 28% with a P value of 1.08 × 10−41 (Kathiresan et al. 2009). In another study published by CARDIoGRAM consortium with genotypes of more than 60,000 individuals in 2011, rs4977574 was found to be associated with coronary artery diseases with a p value of 1.35 × 10−22 and the frequency of rs4977574 was reported to be 46% (Schunkert et al. 2011). CDKN2B is a gene located ~89 kilobases from rs4977574. rs4977574 was found to be associated with the increased mRNA level of CDKN2B in human fat tissue and therefore, rs4977574 might have a role in the onset of MI by leading to atherosclerosis through CDKN2B gene (Kathiresan et al. 2009). The genomic region that includes rs4977574 and other SNPs associated with coronary artery diseases has been shown to regulate the expression of CDKN2A and CDKN2B genes in a mouse model. In the same mouse model, aortic smooth muscle cells exhibited excessive proliferation and impaired cell senescence (Visel et al. 2010). Overall, rs4977574 may predispose individuals for MI by affecting the expression of nearby genes and the proliferation capacity of cells. These findings may give a mechanistic explanation for the relationship between rs4977574 and MI.
A novel PCR-RFLP was developed to reveal the genotype for rs4977574 in patients with MI and controls. To the best of our knowledge, there is not a published PCR-RFLP protocol developed for the analysis of rs4977574. Using this PCR-RFLP method, it is possible to study a higher number of patients with MI to reveal the presence of rs4977574 in a cost-effective manner. The frequency of rs4977574 was shown to be higher in patients with MI compared to controls in Turkish population in this pilot study. The clinical significance and association of this SNP needs to be further studied in the pathogenic progression of MI using a higher number of patients. rs4977574 is in the close proximity of CDKN2A, CDKN2B and ANRIL. Recently, it has been reported that expression levels of CDKN2A, CDKN2B and ANRIL may be associated with atherosclerosis and cardiovascular disease (Congrains et al. 2012; Motterle et al. 2012). Therefore, rs4977574 may have clinical implications including atherosclerosis and abnormal smooth muscle proliferation in coronary arteries.
In further studies, it will be possible to investigate the frequency and the pathogenic significance of rs4977574 in subgroups of patients with MI. In addition, the expression of CDKN2B gene can be assessed in patients carrying rs4977574 allele and in patients not carrying the rs4977574 allele. More importantly, a large group of patients with MI can be genotyped for the presence of rs4977574, and these patients can be followed for long term to find out associations with clinical outcomes, progression of MI, atherosclerosis and rs4977574 genotype.
In this study, for the first time, a new PCR-RFLP method was designed to identify the occurrence of the SNP, rs4977574. This PCR-RFLP method was applied for the first time and an increased frequency of rs4977574 was demonstrated in patients with MI in Turkish population.
This work was supported by Erciyes University Scientific Research Fund (EU-BAP), Grant number: CAP-12-4044 (C. Sakalar).
All of the authors declare that they have no conflict of interest and no financial relationship with any commercial entity that has an interest in the subject of this manuscript.