2013 Volume 231 Issue 2 Pages 127-138
2013 Volume 231 Issue 2 Pages 127-138
Autophagy is a protective cellular mechanism in response to various stresses, including sepsis. Sepsis is defined as systemic inflammation by infection. Surfactant protein A and D (SP-A and SP-D) are involved in host defense, regulation of inflammation, and homeostasis, but their roles in the autophagic activity and relevant gene expression in sepsis are unclear. In this study, mice lacking SP-A and SP-D (SP-A/D KO mice) and background-matched wild-type (WT) C57BL/6 mice underwent either cecal ligation and puncture (CLP) or sham surgery. The results showed that SP-A/D KO mice had lower mortality than WT mice in CLP sepsis. Liver tissues showed marked pathological changes in both septic SP-A/D KO and WT mice 24 hrs after CLP treatment; and quantitative analysis of liver histopathology revealed significant difference between septic SP-A/D and septic WT mice. SP-A/D KO mice had higher basal and sepsis-induced level of autophagy than WT mice (p < 0.05), as judged by Western blot and electron microscopic analyses. The expression of 84 autophagy-related genes revealed differential basal and sepsis-induced gene expression between SP-A/D KO and WT mice. The expression increased in three genes and decreased in four genes in septic WT mice, as compared to septic SP-A/D KO mice (p < 0.05). Furthermore, differential responses to sepsis between SP-A/D KO and WT mice were found in six signaling pathways related to autophagy and apoptosis. Therefore, enhanced autophagic activity improves the survival of septic SP-A/D KO mice through the regulation of liver autophagy/apoptosis-related gene expression and signaling pathway activation.
Sepsis is defined as systemic signs of inflammation (fever, leukocytosis, tachycardia and tachypnea) to a proven or suspected infectious cause (Anthony Fauci 2011). In the United States, there are approximately 750,000 cases of sepsis per year, resulting in about 250,000 deaths annually (Angus et al. 2001; Martin et al. 2003), which underscores the need to gain an understanding of the pathophysiology of cellular and molecular mechanisms involved in sepsis.
The liver is a primary organ in the response to sepsis and plays both victim and actor roles in the pathogenesis of sepsis. Primary hepatic dysfunction is a reversible injury that can be detected in the first hours following initial injury. This usually occurs as a result of hypoperfusion and can lead to disseminated intravascular coagulation. Secondary hepatic dysfunction is silent and accounts for a spillover of bacteria, endotoxin and inflammatory mediators. The uptake and detoxification of bacterial products activates inflammatory pathways in hepatic Kupffer cells (Cerra et al. 1979; Dhainaut et al. 2001) which may promote multiple organ dysfunction along the gut-liver axis (Knaus et al. 1985; Carrico et al. 1986; Dhainaut et al. 2001).
Autophagy is the major intracellular degradation mechanism by which cytoplasmic proteins and organelles are delivered to and degraded in the lysosome (Mizushima and Komatsu 2011). It contributes to cellular survival during nutrient deprivation and turnover of damaged organelles. Autophagy can be induced by extracellular and intracellular signals, including oxidative stress, endoplasmic reticulum (ER) stresses, glucose, amino acids, serum starvation and sepsis (Levine and Kroemer 2008; Mizushima et al. 2008; Eskelinen and Saftig 2009; Mizushima 2009; Munz 2009; Tanida 2011). It is a multistep process and the intermediate double membrane vesicle or “autophagosome” has a key role in transporting degraded cytoplasmic materials to the lysosome (Mizushima and Komatsu 2011). Previous studies have shown a protective role for autophagy by limiting cell death, apoptosis and irreversible organ dysfunction (Levine and Yuan 2005; Carchman et al. 2011; Nakahira et al. 2011). Recently, it has been observed that autophagic activity increased in hepatocytes in septic patients and mouse models (Watanabe et al. 2009) and septic rat model (Chien et al. 2011). Liver is a primary organ to response to cecal ligation and puncture (CLP) sepsis at the early stage and autophagic activity in the liver exhibited remarkable changes in the first hours after CLP surgery (Chien et al. 2011). Therefore, we chose liver tissue to investigate changes of autophagic activity and autophagy-related gene expression in the present study.
Surfactant protein A and D (SP-A and SP-D) are members of the C-type lectin (collectin) family and are part of the innate immune system. They play an important role in host defense and the regulation of inflammation in various infections (Haagsman et al. 2008). Although the lung is the main site of SP-A and SP-D expression, their expression was also found in several other organs such as trachea, brain, testis, salivary gland, heart, prostate gland, kidney, and pancreas (Madsen et al. 2000). The SP-A and SP-D proteins consist of 4 domains: 1) N-terminal region, 2) triple helical collagen-like domain, 3) neck region, and 4) carbohydrate recognition domain (CRD) (Seaton et al. 2010). In healthy mammals, SP-A and SP-D bind to a signal regulatory protein-a (SIRP-a) of the macrophage by their CRD domain and prevent expression of inflammatory mediators (Gardai et al. 2003). However, in the presence of microbes, CRD interacts with carbohydrate molecules on the surface of the microbes (Gardai et al. 2003), the collagenous tail binds to CD91 of macrophages and stimulates Nf-κB activation, which leads to the production of inflammatory mediators (Gardai et al. 2003).
In the present study, we investigated autophagic activity and relevant gene expression in the pathogenesis of CLP sepsis with SP-A and SP-D double knockout (SP-A/D KO) and background-matched C57BL/6 wild type (WT) male mice. We observed different survival rates between SP-A/D KO and WT mice following CLP treatment. We found differential liver function, autophagic activity and autophagy-related gene expression patterns, as well as associated signaling pathways in the livers of septic SP-A/D KO and WT mice. These findings suggest SP-A and SP-D play a role in the regulation of autophagic activity and relevant signal pathways in the pathogenesis of sepsis.
Male mice (8-12 weeks old) were used in this study. The original SP-A/D KO mice with a C57BL/6 background were kindly provided by Dr. Hawgood of The University of California San Francisco (Hawgood et al. 2002). The SP-A/D KO mice had been backcrossed at least 10 generations with C57BL/6 mice and there are no differences in body weights between SP-A/D KO and matched WT mice. After both SP-A and SP-D genes were disrupted no SP-A and SP-D protein was expressed in all tissues of the SP-A/D KO mice (Hawgood et al. 2002). All SP-A/D KO mice were bred in the animal core facility at SUNY Upstate Medical University under pathogen-free conditions. Background and age-matched WT C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME) or bred in our animal core facility. All animal experiments were conducted in accordance with the Institutional Animal Care and Use Committee guidelines of SUNY Upstate Medical University (Protocol # CHUA 270) and the National Institutes of Health guidelines on the use of laboratory animals.
Cecal ligation and puncture (CLP) operationPolymicrobial sepsis was induced by subjecting mice to CLP operation (Rittirsch et al. 2009). Male SP-A/D KO and WT mice were acclimated for 1 week with free access to food and water. Prior to surgery, the mice were anesthetized using intraperitoneal ketamine/xylazine (90 mg/kg ketamine, 10 mg/kg xylazine). After laparotomy, the cecum was ligated at about 1.0 cm position from distal pole to the base of the cecum with a 5-0 silk suture, the ligature did not interrupt bowel continuity, the cecum was punctured twice with a 22-gauge needle and gently squeezed to extrude a 1-mm3 column of fecal material (in CLP mice), and then the cecum was returned to the abdominal cavity. In sham operation, the cecum was located and manipulated but neither ligated nor punctured, and then the abdominal incision was closed with 5-0 silk suture. After CLP, the mice were resuscitated with 1 ml warmed sterile saline injected subcutaneously. Buprenorphin (0.05 mg/kg body weight) was injected for postoperative analgesia every 12 hrs. For survival study, the animals were monitored for 48 hrs or 7 days, respectively, in two independent experiments. For all other studies, mice were sacrificed under general anesthesia at 6 or 24 hrs.
Collection of mouse liver tissuesMice were anesthetized with interperitoneal ketamine/xylazine (90 mg/kg ketamine, and 10 mg/kg xylazine) and then the abdomen of the animals was opened via a midline incision and exsanguinated by transecting the vena and aorta. Liver tissues were excised and either immediately frozen in liquid nitrogen or fixed in 10% neutral formalin for subsequent histological analysis, or in 2.5% paraformaldehyde and 4% glutaraldehyde for electron microscopy analysis (see below). Sections from five mice for each condition were stained with hematoxylin and eosin (H+E) and then assessed qualitatively for sepsis-related histopathology by two experienced investigators.
Biochemical analysis of liver functionsBlood samples were collected from tail vein at 6 and 24 hrs after CLP and control mice. The levels of serum aspartate aminotransferase (AST, also called serum glutamate-oxaloacetate transferase (SGOT)), and Alanine aminotransferase (ALT, also known as serum glutamic-pyruvic transferase (SGPT)) were measured to monitor the liver function by colorimetric kits (Sigma-Aldrich, St. Louis). Levels of AST and ALT were determined according to the manufacturer’s protocol. In brief, serum sample (20 μl/well) was added into the Master Reaction solution and then incubated at 37°C for 30 min (ALT) or 60 min (AST). The absorbance of the solution was determined at 570 nm for ALT or at 450 nm for AST using a microplate reader (Multiskan Ascent, Thermo Labsystems).
Electron microscopic analysis of hepatocytes and determination of autophagosome numberLiver tissues were used for transmission electron microscopy after 24 hrs of fixation with 2.5% paraformaldehyde and 4% glutaraldehyde as described previously (Wang et al. 2010). Specimens were stained with 1% osmium tetroxide and 1.5% potassium ferrocyanide, and subsequently embedded in Emed812 resin (Electron Microscopy Science). Ultrathin sections (90 nm) were prepared and stained with 2% aqueous uranyl acetate and lead citrate for electron microscopic analysis. Several sections from each of the specimens were examined at the range from 10,000 × to 40,000 × magnification. To study the formation of autophagsomes we have examined at least 100 fields at the 40,000 × magnification for each specimen. Total 20 photographs were taken from randomly selected grids from each section by a random number generator, and the numbers of autophagosomes (autophagic vacuoles) were recorded for each photograph.
Gene expression analysis by RT2 Profiler PCR Array SystemThe expression of 84 autophagy-related genes in the liver of mice was examined using the RT2 Profiler PCR array kit (Cat. No. PAMM-084Z, SuperArray Bioscience, Frederick, MD). Total RNA was isolated from the liver homogenates using the RNA-Bee reagent (Tel-test, Friendswood, TX) according to manufacturer’s instructions. RNA concentrations were determined by spectrophotometry. cDNA was synthesized from 1 µg of total RNA using RT² First Strand Kit (SuperArray Bioscience, Frederick, MD). Real-time PCR was performed with the RT2 Profiler PCR array system according to manufacturer’s instructions using ABI Step-One (Applied Biosystems, Foster City, CA). The expression levels of the mRNA of each gene in different groups were normalized using the expression of housekeeping genes (Gusb, Hprt, Hsp90ab1, Gapdh, and Actb). Relative gene expression changes were calculated using the 2−(averageΔΔCt) methods (White et al. 2012). Fold change (FC) values are presented for genes treated relative to control samples. Significant changes in gene expression between groups were identified and analyzed (p < 0.05 by t-test and FC ≥ 1.5). Level of significance (FC ≥ 1.5) was determined as described previously (White et al. 2012).
Signaling pathway association analysisSignaling pathways associated with genes were analyzed with the ingenuity pathway analysis program (ingenuity system, Redwood City, CA), which can gain additional insight into functional significance and signaling pathway association in response to sepsis.
Western blot analysis for protein expression in liverLiver tissues were homogenized in RIPA buffer containing cocktails of protease inhibitors (Roche Molecular Biochemicals, IN). The samples were centrifuged at 13,000 × g for 15 min and supernatant was recovered for the analysis that follows. Protein concentrations were determined using the micro-BCA protein assay kit (Pierce Biochemicals, FL). A total of 15 µg of proteins were subjected separation by electrophoresis on 12% SDS-PAGE gel, and then transferred to PVDF membrane as described previously (Wang et al. 2003). The membrane was probed with LC3-II monoclonal antibody (Cat. No. 3868, Cell Signaling Tech. INC., Danvers, MA), β-actin antibody (Santa Cruz Biotechnology, CA), or Caspase-3 (Santa Cruz Biotechnology, CA), and subsequently incubated with a secondary HRP-conjugated antibody (Bio-Rad, Hercules, CA). Bands were detected using ECL Western Blotting Detection Reagent (Pierce Biochemicals, FL) and the blots were exposed to X-film (Pierce Biochemicals, FL). The bands on the films were quantified by the Quantity One software (Bio-Rad, CA, USA).
Statistical analysisData were expressed as means ± SE. Statistical analyses of the data were carried out using SigmaStat version 3.0 (Jandel Scientific, CA). Differences between and among groups were assessed by Student’s t-test or one-way ANOVA test. Animal survival experiments were assessed by a Kaplan-Meier survival analysis. P < 0.05 was considered as statistically significant.
We compared survival rates of SP-A/D KO and WT mice (n = 25 mice per group) after CLP surgery in two independent experiments, in which survival of animals for each experiment was monitored for 48 hrs or 7 days, respectively. The results from both experiments indicated that the survival rate of septic WT mice was significantly lower than that of septic SP-A/D KO mice (p < 0.05). In the 48-hrs survival study, survival rates of septic WT and SP-A/D KO mice were about 25% and 50% 48 hrs post CLP surgery (Fig. 1). In the 7-days survival study, we found 12% and 50% survival of septic WT and SP-A/D KO mice, respectively, by 7 days after CLP surgery. The results of both survival experiments are identical. Furthermore, we compared the survival rate of septic WT mice grown in two different facilities, i.e. either purchased from the Jackson Laboratories or bred in our animal core facility. The results indicated no difference of the survival rate of these WT mice grown in two facilities in this study. Therefore, WT mice from the Jackson Laboratories were used in following studies.

Difference of survival rates between SP-A/D KO and WT mice after CLP surgery.
SP-A/D KO and WT mice were subjected to CLP surgery, and survival (n = 25 for each CLP group, and n = 10 for each sham group) was monitored for 48 hours. Differential survival rates were observed in septic SP-A/D KO and WT mice, i.e. higher survival rate in septic SP-A/D KO mice compared to septic WT mice (*p < 0.05).
Since the liver is a primary organ in the response to septic injury, we examined histopathological changes in septic SP-A/D KO and WT mice, as well as in control (sham) mice. The results showed obvious pathological changes in the septic SP-A/D KO and WT mice 24 hrs after CLP surgery, but not in the sham mice (Fig. 2A). The hepatocytes in CLP mice showed heavy vacuolation and cytoplasmic degradation, loss of discernible cell junctions, and in extreme instances the nuclei were missing. The histopathological assessment of the livers was consistent with mouse sepsis in this study. Quantitative analysis of histopathological scores revealed significant difference between septic SP-A/D KO and septic WT mice (Fig. 2B). Furthermore, we examined liver function by analyzing serum AST and ALT levels in the septic SP-A/D KO and WT mice, as well as sham mice. The results showed that the levels of both AST and ALT increased significantly in the septic SP-A/D KO and WT mice 24 hrs after CLP surgery compared to sham mice (AST: septic SP-A/D KO vs. sham SP-A/D KO mice = 320 ± 33 vs. 125 ± 26 mg/dl, p < 0.01, septic WT vs. sham WT mice = 423 ± 52 vs. 135 ±21 mg/dl, p < 0.01; ALT: septic SP-A/D KO vs. sham SP-A/D KO mice = 274 ± 23 vs. 89 ± 16 mg/dl, p < 0.01, septic WT vs. sham WT mice = 356 ± 32 vs. 94 ± 17 mg/dl, p < 0.01). The levels of AST and ALT septic SP-A/D KO mice were lower (p < 0.05) than that of septic WT mice 24 hrs after CLP surgery. These results indicate that septic SP-A/D KO mice had better liver function 24 hrs after CLP surgery.
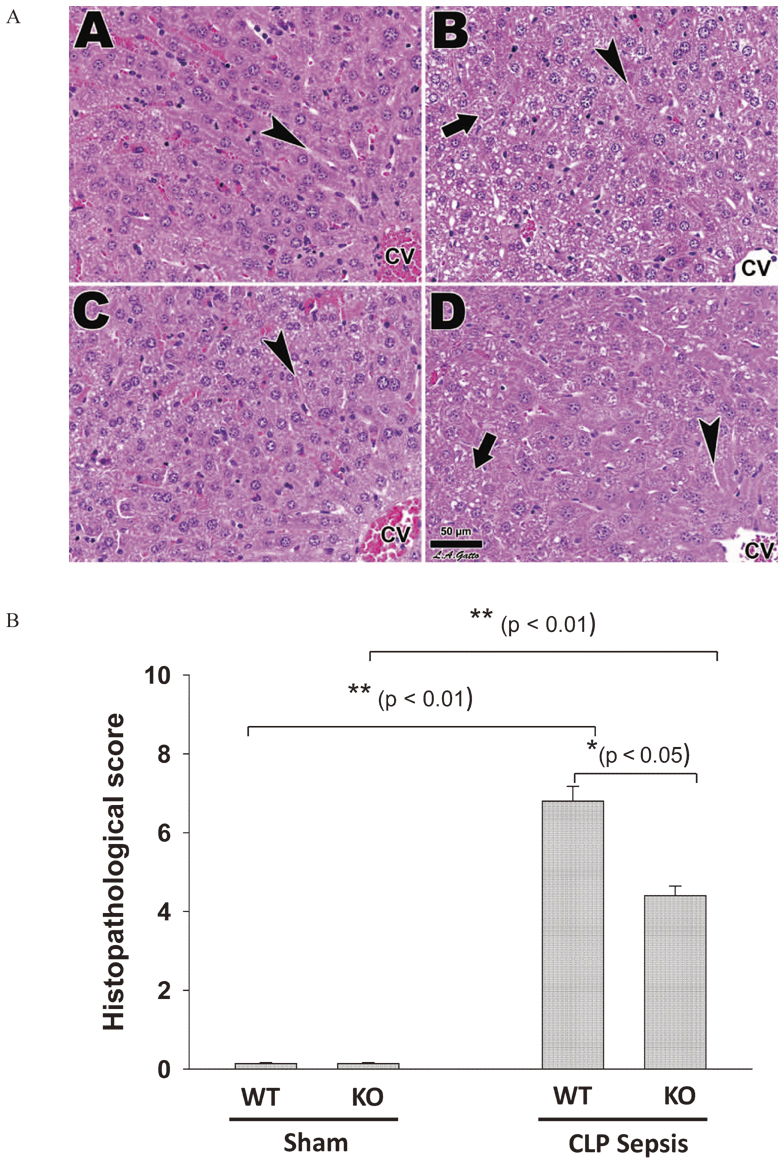
Histology of the liver in SP-A/D KO and WT mice after CLP surgery.
Fig. 2A shows that Liver histology from WT and SP-A/D KO mice receiving either sham (A and C) or CLP surgery (B and D), respectively. All samples exhibited comparable liver lobule architecture, as liver sinusoids (arrowheads) extended between rows of hepatocytes, and drained into the central vein (CV). The CLP (B and D) were marked by hepatocytes showing heavy vacuolation and cytoplasmic degradation, loss of discernible cell junctions, and in extreme instances the nucleus was missing (arrows). Fig. 2B shows quantitative difference of histopathological scores between septic WT and septic SP-A/D KO mice (p < 0.05, n = 5 mice/group).
To assess autophagic activity in the livers of SP-A/D KO and WT mice 6 hrs and 24 hrs after CLP and sham surgery, we examined expression of LC3-II (Microtubule associated protein 1, Light chain 3, in its conjugated form) by Western blot analysis (Fig. 3A). Because LC3-II is widely used as a biomarker of autophagic activity, higher level of LC3-II expression represents higher autophagic activity in the tissue. For standardization of LC3-II expression, the blot was stripped and reprobed with antibody to β-actin. The ratio of LC3-II to β-actin level represents autophagic activity of each sample. The results (Fig. 3A and B) demonstrated: a) The basal level of autophagic activity was higher in SP-A/D KO mice compared to in WT mice (p < 0.01); b) 6 hrs and 24 hrs after CLP surgery, the level of LC3-II in the livers of both septic SP-A/D KO and WT mice increased significantly compared to sham mice (p < 0.01), and c) a higher level of LC3-II was maintained for 24 hrs in both septic SP-A/D KO and WT mice.

Autophagic activity in the livers of SP-A/D KO and WT mice after CLP surgery.
Analysis of autophagic activity in the livers by determining the level of protein LC3-II which is used as a biomarker of autophagic activity.
Liver tissues were harvested at 6 hrs and 24 hrs after CLP or sham surgery. Total proteins were subjected separation by electrophoresis and the autophagic activity marker LC3-II and β-actin were detected by western blot analysis (Panel A) and the blots were quantified (Panel B). The basal level of autophagic activity was higher in the sham SP-A/D KO mice than in sham WT mice (p < 0.05). Autophagic activity was increased in both septic SP-A/D KO and WT mice from 6-24 hrs after CLP (p < 0.01, CLP SP-A/D KO vs. sham SP-A/D KO, or CLP WT vs. sham WT) and difference was observed between septic SP-A/D KO vs. septic WT mice at 6 hrs (Panel B) and 24 hrs (data not shown) after CLP (n = 5 mice/group).
To further confirm the formation of autophagosomes in the hepatocytes, liver tissues of SP-A/D KO and WT mice were examined by transmission electron microscopy (Fig. 4A. A = WT/Sham, B = WT/CLP, C = KO/Sham, D = KO/CLP). Autophagosomes (autophagic vacuoles) in five sections of each specimen from CLP or sham surgery were counted in each section for 20 random fields (40,000 ×). The results in Fig. 4B from 6 hrs after CLP and sham surgery showed: a) the number of autophagosomes was larger in the sham SP-A/D KO mice than in sham WT (p < 0.01), suggesting higher basal levels of autophagic activity in the SP-A/D KO mice than in the WT mice. b) the number of autophagosomes increased significantly in septic SP-A/D KO and WT mice compared to sham SP-A/D KO and WT mice (p < 0.01); c) Septic SP-A/D KO mice exhibited more autophagosomes than septic WT mice (p < 0.01). Similar results were observed 24 hrs after CLP treatment (data not shown). These observations from electron microscopic analysis were consistent with those from autophagic biomarker LC3-II activity.

Analysis of autophygosomes in the livers of SP-A/D KO and WT mice by transmission electron microscopy.
Liver tissues were harvested from SP-A/D KO and WT mice in 6 hrs after CLP or sham surgery. Ultrastructure was examined in at least five liver tissue sections per group by transmission electron microscopy (Panel A: A = WT/Sham, B = WT/CLP, C = KO/Sham, D = KO/CLP). Autophagosomes in the liver cells are marked by arrows (Panel A). The numbers of autophagosomes were counted in 20 random fields under electron microscopy (Panel B). The results indicated higher numbers of autophagosomes in SP-A/D KO mice (p < 0.01, sham SP-A/D KO vs. sham WT, or septic SP-A/D KO vs. septic WT). At 6 hrs after CLP septic SP-A/D KO and WT mice have significantly higher numbers of autophagosomes compared to sham SP-A/D KO and WT mice, respectively (p < 0.01). Septic SP-A/D KO mice exhibited higher numbers of autophagosomes than septic WT mice (p < 0.01).
To explore molecular mechanisms underlying the difference in autophagic activity between SP-A/D KO and WT mice, we examined mRNA levels of 84 autophagy-related gene expressions in the liver tissues of SP-A/D KO and WT mice 6 hrs after CLP and sham surgery. The mRNA levels of 84 genes of four mouse groups, i.e. sham SP-A/D KO and WT mice, and septic SP-A/D KO and WT mice, were determined by real-time PCR Array system as described in the section of methods. The relative levels of each gene expression were compared among four groups: sham WT vs. sham SP-A/D KO, septic SP-A/D KO vs. sham SP-A/D KO, septic WT vs. sham WT, septic WT vs. septic SP-A/D KO. The genes with significant changes (FC value > 1.5 and p < 0.05) were showed in the Fig. 5 and Table 1.
a) Comparison between sham WT and sham SP-A/D KO mice: Fig. 5A shows that the mRNA levels of genes Bid, Ctsb and Rps6kb1 expression were higher (FC > 2) in WT mice than in SP-A/D KO mice, but the mRNA levels of Atg9a and Bak-1 expression were higher (FC > 2) in SP-A/D KO mice than in WT mice.
b) Comparison between septic SP-A/D KO and sham SP-A/D KO mice: The expression of 4 genes (Atg16l2, Ctsb, Rps6kb1 and Tgm2) increased significantly (> 2 fold) in the septic SP-A/D KO compared to sham K/O mice, but the expression of gene Cxcr4 decreased (> 2 times) in the septic SP-A/D KO compared to sham SP-A/D K/O mice (Fig. 5B).
c) Comparison between septic WT and sham WT mice: Only moderate increases of 6 gene (Atg7, Irgm1, Map1lc3b, Pten, Sqstm1) expressions were observed in septic WT compared to sham WT mice (Fig. 5C). The moderate increases in autophagy-related genes indicated that autophagy was occurring in septic WT mice but with lesser magnificence than in septic SP-A/D KO mice.
d) Comparison between septic WT and septic SP-A/D KO mice: The levels of three gene (Arsa, Hgs, FADD) expressions were significantly higher in septic WT mice compared to septic SP-A/D KO, but 4 genes (Bak-1, Nf-κB, p53, and Atg12) had lower levels of expression in septic WT compared to the septic SP-A/D KO mice. These were suggestive of more dominant autophagic activity in septic SP-A/D KO mice compared to septic WT mice.

Differential autophagy-related gene expression in the liver of SP-A/D KO and WT mice after CLP surgery.
Liver tissues of septic and sham SP-A/D KO and WT mice were harvested 6 hrs after CLP surgery. Total RNA was isolated from the liver of each mouse by using the RNA-Bee reagents. The mRNA levels of the 84 autophagy-related gene expression in four mouse groups were analyzed by RT2 Profile PCR Array system. To assess the difference of mRNA expression of each gene between two groups, the fold change (FC) values of average mRNA level for each gene were calculated relative to control samples. The FC values represent the difference between two groups, i.e. sham WT vs. sham SP-A/D KO (Panel A); septic SP-A/D KO vs. sham SP-A/D KO (Panel B); septic WT vs. sham WT (Panel C); septic WT vs. septic SP-A/D KO (Panel D).

Differentially expressed genes in septic mice.
To gain further insight into the signaling pathways involved in septic SP-A/D KO and WT mice, those genes with significant expression (FC > 1.5 and p < 0.05) between septic SP-A/D KO and septic WT mice were used for analysis with the ingenuity pathway analysis program. Six signaling pathways related to autophagy and apoptosis were identified to be associated with the different response to sepsis between SP-A/D KO and WT mice (Fig. 6A). They included: 1) pathway of the role of protein kinase receptor (PKR) in interferon induction and antiviral response; 2) TNF-like weak inducer of apoptosis (TWEAK) signaling pathway; 3) apoptosis signaling pathway; 4) TNF receptor 1 (TNFR1) signaling pathway; 5) Myc-mediated apoptosis signaling pathway; 6) death receptor signaling pathway. As an example, Figure 6B shows the TNFR1 signaling pathway, in which the expression of 5 identified genes was up-regulated in the septic SP-A/D KO mice, compared to the septic WT mice. In order to confirm difference of autophagy and apoptosis-gene expression, we further examined activated caspase-3 (a biomarker related to apoptosis) in the liver tissues of septic mice (24 hrs after CLP surgery) we observed significant increase (p < 0.01) of activated caspase-3 in the livers of septic mice compared to sham mice and the level of activated caspase-3 in the lever of septic WT mice was higher (p < 0.05) than that in septic SP-A/D KO mice (Fig. 6C). The differences of signaling pathways and relative gene expression in septic SP-A/D KO and WT mice provided with evidence that the SP-A and/or SP-D could regulate or influence the expression of other genes involved in autophagy and apoptosis-related signaling pathways in sepsis.

Signaling pathways associated with differential response to sepsis between septic SP-A/D KO and WT mice.
Six related signaling pathways were identified to be associated with differential response to CLP treatment between septic SP-A/D KO and WT mice (Fig. 6A). One representative TNFR1 signaling pathway is shown here in which five key components (green) of the pathway showed upregulated levels in septic SP-A/D KO mice compared to septic WT mice (Fig. 6B). One representative apoptosis biomarker (caspase-3) expression was examined by Western blot analysis in the liver tissues of SP-A/D KO and WT mice. The level of activated caspase-3 in the liver tissues of septic WT mice was higher than that of septic SP-A/D KO mice (p < 0.05) (Fig. 6C).
SP-A and SP-D proteins are part of the innate immune system, and their protective role in respiratory infections has been studied extensively (Wright 2005; Haagsman et al. 2008). However, the effects of SP-A and SP-D in the pathogenesis of sepsis and the regulation of autophagy are unknown. In the present study we found that SP-A/D KO mice had higher basal and sepsis-induced levels of autophagic activity and higher survival rates compared to septic WT mice. Analysis of autophagy-related gene expression revealed differential gene expression patterns between SP-A/D KO and WT mice with and without CLP surgery. These findings suggest that SP-A and SP-D are involved in the regulation of autophagy activity during abdominal sepsis.
LC3-II is usually chosen as a biomarker of autophagic activity because it is tightly bound to the autophagosomal membrane. LC3 proteins, consisting of 7 members, are all key components of the autophagy machinery. They are required for elongation and closure of the autophagosome (Puissant et al. 2012). In the present study we observed higher basal and sepsis-induced levels of autophagic activity in the livers of SP-A/D KO mice compared to WT mice, by Western blot analysis of both LC3-II protein and the counts of autophagosome numbers in hepatocytes under electron microscopy. A recent report using rat CLP-induced sepsis model showed increased autophagic activity in the hepatocytes 6 hrs after septic surgery but decreased level by 18 hrs post surgery (Chien et al. 2011). In contrast, our mouse sepsis model did not show decrease in autophagic activity by 24 hrs after CLP surgery. The difference may reflect difference between species. Similarly, Watanabe et al. observed increased numbers of autophagosomes in the livers of septic patients and CLP mice but failed to obtain evidence of increased autophagy-related gene expression in response to sepsis and therefore could not conclude whether an increase in autophagosome numbers had a beneficial role in response to sepsis (Watanabe et al. 2009). In the present study we observed increased autophagic activity by LC3-II biomarker and the ultrastructural analysis of hepatocytes after CLP surgery, and the increased autophagy-related gene expression was consistent with observed improvement in survival from sepsis. Our findings suggest collectively that autophagy is a protective mechanism in response to sepsis, as the higher autophagic activity is associated with the lower mortality. Indeed, we have observed higher survival rates in septic SP-A/D KO mice compared to septic WT mice.
Sepsis initiates complex immunologic responses that vary over time. The magnitude of the inflammatory response to sepsis varies depending on many factors (Hotchkiss et al. 2009). Both apoptotic and autophagy pathways may be activated in response to septic insults (Ayala et al. 2008; Hotchkiss et al. 2009; Choi and Ryter 2011). They share many genes and pathways, as well as their molecular regulators (Eisenberg-Lerner et al. 2009). But there are some genes that are more specific to either pathway, for instance, FADD (FAS-Associating Protein with Death Domain), Bak-1(Bcl-2 Antagonist Killer-1) and Bid (BH3-Iinteracting Domain Death Agonist) are well known pro-apoptotic proteins (Li et al. 1998; Yeh et al. 1998; Borner 2003; Yeretssian et al. 2011). Atg proteins (autophagy related genes with a total of 30 members), on the other hand, are more specific to autophagy and play essential roles in the formation and elongation of the phagophore (Puissant et al. 2012). In the present study we found an increased expression of the two autophagy related genes (Atg16l2 and Atg7) in response to sepsis in SP-A/D KO and WT mice, respectively (Fig. 5B and C). Furthermore, we observed increased expression of Atg12 gene in septic SP-A/D KO mice as compared to septic WT mice (Fig. 5D). The results of the autophagy-related gene expression are consistent with the observation of autophagic activity in septic SP-A/D KO and WT mice. Of interest, we found that SP-A/D KO mice had increased expression of Atg9 and higher basal level of autophagic activity compared to WT mice, suggesting potential regulatory functions for SP-A and SP-D in autophagic activity. Dual immune modulatory functions of SP-A and SP-D have been reported (Gardai et al. 2003). When comparing mice of same genotype with or without sepsis, the autophagy-related gene Tgm-2 except for Atg genes showed increased expression in septic SP-A/D KO vs. sham SP-A/D KO mice (Fig. 5B), and Irgm1, Sqstm-1 and Map1lc3b expressed more in septic WT compared to sham WT mice (Fig. 5C), indicating a sepsis-induced increase in autophagic activation.
An exaggerated inflammatory response to infection by the host contributes to dysfunction of vital organs such as lung, liver, kidney and heart, which is the major cause of morbidity and mortality in sepsis. Multiple cell populations and cell signaling pathways are involved and interact in these complex processes, especially the PI3K/Akt and Nf-ĸB pathways (Abraham and Singer 2007; Manukyan et al. 2010). Increased levels of NF-κB expression were observed in the septic SP-A/D KO mice in the present study. The relationship between NF-κB and autophagy has been extensively studied (Djavaheri-Mergny et al. 2006). In cells lacking NF-κB activation, TNF-α upregulates the expression of the autophagy-promoting protein Beclin-1 and subsequently induces the accumulation of autophagic vacuoles (Djavaheri-Mergny et al. 2006). SP-A is involved in the regulation of NF-κB signaling pathway and SP-A can downregulate NF-κB activation (Wu et al. 2003). This observation explains the increased level of Nf-κB and higher survival rates in septic SP-A/D KO mice compared to septic WT mice.
In summary, the present study examines autophagic activity and autophagy-related gene expression in the livers of SP-A/D KO and WT mice in CLP sepsis model. Our findings suggest that autophagy has a protective effect in response to sepsis and that SP-A and SP-D are involved in the regulation of autophagic activation. Enhanced basal and sepsis-induced autophagic activity in SP-A/D KO mice significantly improved liver function and survival rate compared to in WT mice. Analysis of the expression of 84 autophagy-related genes demonstrated differential basal and sepsis-induced expression, involved in autophagy and apoptosis, between septic SP-A/D and WT mice. Furthermore, six signaling pathways related to autophagy and apoptosis were identified to be associated with differential responses to sepsis when comparing between septic SP-A/D KO and WT mice. Further investigations are necessary to gain a better understanding of the molecular mechanisms underlying the regulatory role of SP-A and SP-D on autophagy and apoptosis in sepsis.
We would like to thank Dr. S. Hawgood of The University of California San Francisco, CA for kindly providing SP-A/D KO mice. This work was in part supported by NIH grant HL096007 and the Research Fund from SUNY Upstate Medical University.
The authors declare no conflict of interest.