2017 Volume 241 Issue 4 Pages 297-308
2017 Volume 241 Issue 4 Pages 297-308
Coronary artery disease (CAD) is a multifactorial disease with a genetic component. Pigment epithelium-derived factor (PEDF) exerts anti-inflammatory, anti-oxidant, anti-thrombotic, and anti-angiogenic effects and thus has received increasing attention as a sensitive biomarker of atherosclerosis and CAD. To explore the potential association between PEDF single nucleotide polymorphisms (SNPs) and CAD, we performed this case-control study of consecutive elderly Chinese Han male patients (n = 416) and age-matched male controls (n = 528) without a history of CAD or electrocardiographic signs of CAD. The enrolled CAD patients (age ≥ 60 years) are not biologically related. A tag approach was used to examine 100% of common variations in the PEDF gene (r2 ≥ 0.8, minor allele frequency > 0.1). PEDF tag SNPs (tSNPs) were selected using the HapMap Data-CHB which describes the common patterns of human DNA sequence variation and Tagger program. SNPs were genotyped using ligase detection reaction (LDR). Seven tSNPs (rs8075977, rs11658342, rs1136287, rs12603825, rs12453107, rs6828 and rs11078634) were selected. Among them, only one SNP, rs8075977 (C/T) located in the 5'-flanking region, showed the significant effect on the susceptibility to CAD. The frequency of its T allele was significantly higher in the controls (52.7%) than that in the CAD group (46.2%) (adjusted OR = 0.88, 95% CI: 0.80-0.96; P = 0.005). In conclusion, the T allele of rs8075977 in the 5'-flanking region of the PEDF gene may be protective for CAD. Conversely, the C allele at this variation site is associated with CAD in elderly Chinese Han men.
Coronary artery disease (CAD) is a common cause of morbidity and mortality worldwide (GBD 2013 Mortality and Causes of Death Collaborators 2015). Atherosclerosis, a chronic inflammatory disorder, is believed to be the chief cause of CAD (Li 2011). Many risk factors have been identified for CAD, including hypertension, diabetes, dyslipidemia, smoking and obesity. In addition to these traditional risk factors, genetic factors that are predisposing to atherosclerosis have also attained considerable interest (Roberts and Stewart 2012; Yuan et al. 2014), and growing studies have attempted to identify genetic risk factors for CAD. It is reported that premature CAD is closely associated with specific family history (Yucel et al. 2012). A close relative with CAD increases the risk of developing CAD (Mayer et al. 2007; Haroon et al. 2015). Moreover, a mounting number of susceptibility genes linked to CAD have been reported (O’Donnell et al. 2011; Schunkert et al. 2011). Recent studies have established a role for single nucleotide polymorphisms (SNPs) in CAD development or prognosis, including the histone-lysine N-methyltransferase mixed lineage leukemia 5 (MLL5) rs12671368 (G/A) and rs2192932 (G/A) polymorphism, the hepatocyte nuclear factor 1 alpha (HNF1A) rs7310409 (G/A) functional polymorphism and the methylenetetrahydrofolate reductase (MTHFR) rs1801133 (C/T) polymorphism (Chen et al. 2014; Liu et al. 2014d; Yuan et al. 2014). Numerous genome-wide association studies (GWASs) lead to the identification of several novel loci associated with CAD (McPherson et al. 2007; Myocardial Infarction Genetics Consortium 2009; Takeuchi et al. 2012; Ichihara et al. 2013). SNPs associated with CAD are therefore considered to be an important area of study.
Pigment epithelium-derived factor (PEDF), located on chromosome 17p13.3, belongs to the superfamily of serpins (Tombran-Tink et al. 1991; Filleur et al. 2009). It is a multifunctional protein with anti-inflammatory, anti-oxidant, antithrombotic, anti-angiogenic and vasculoprotective properties (Rychli et al. 2009; Yamagishi and Matsui 2014). We previously explored the property of PEDF and have reported that oxidized low-density lipoprotein (ox-LDL) decreases the PEDF expression through inducing reactive oxygen species (ROS) (Liu et al. 2014c), and D-4F, an apolipoprotein A-I mimetic peptide that consists of 4 phenylalanine (F) residues and synthesized from D-amino acids, effectively protects vascular endothelial cells against ox-LDL-induced injury by up-regulation of PEDF (Liu et al. 2014b). Many studies have been performed to investigate the roles of PEDF in CAD and consider PEDF as a sensitive biomarker of the progression of CAD and atherosclerosis. Serum PEDF levels are independently positively correlated with CAD and elevated PEDF may act as a protective response against vascular damage and subsequent CAD (Wang et al. 2013). PEDF processing by matrix metalloproteinase-9 (MMP-9) at the culprit site is associated with coronary plaque rupture (Distelmaier et al. 2012). Our previous results showed that PEDF may be a novel biomarker for the diagnosis and the short-term prognosis in acute coronary syndrome (ACS) (Liu et al. 2014a). Importantly, PEDF has been reported to be a novel biomarker of atherosclerosis in humans (Tahara et al. 2011; Kajikawa et al. 2016). In terms of its function characteristics, PEDF is thought to play a protective role against atherosclerosis by suppressing inflammation, oxidative stress and angiogenesis (Yamagishi et al. 2004; Baba et al. 2005; Maeda et al. 2011; Yamagishi and Matsui 2014). Therefore, PEDF is functionally important in the pathogenesis of CAD and may be a therapeutic target.
PEDF is a reasonable candidate gene for particular diseases in the field of ophthalmology and diabetes, and SNPs of this gene are receiving a great deal of attention. There is evidence suggesting that the PEDF rs12603825 (G/A) is marginally associated with myopic choroidal neovascularization in extremely myopic patients (Miyake et al. 2013). This common functional variant of PEDF gene, rs12603825 (G/A), also contributes to overall body adiposity, obesity-related insulin resistance and circulating leptin levels in humans at increased risk for type 2 diabetes (Bohm et al. 2012). In addition, Met72Thr (rs1136287, C/T), another PEDF gene polymorphism, is associated with an increased risk of wet age-related macular degeneration (AMD) in a Taiwan Chinese cohort (Lin et al. 2008). However, it is unclear whether PEDF SNPs are correlated with CAD. For this reason, we conducted a hospital-based case-control study to genotype a cohort of elderly Chinese Han males including 416 CAD patients and 528 controls. Specifically, the study participants were genotyped for PEDF SNPs to determine whether an association exists between each PEDF SNP and CAD.
Consecutive subjects were recruited from the Chinese PLA General Hospital between October 2009 and May 2010. We enrolled elderly Chinese Han males (age ≥ 60 years) who were not biologically related. The case group included CAD patients who met one of the following criteria: 1) a history of elective or emergency coronary artery bypass grafting (CABG) surgery; 2) a history of elective or emergency percutaneous transluminal coronary angioplasty (PTCA); 3) a history of acute myocardial infarction (ST elevation and Non-ST elevation MI); 4) a history of acute myocardial ischemia but no evidence of infarction (Troponin negative); or 5) ≥ 50% diameter lumen stenosis in a major coronary artery (i.e., left main coronary artery, left anterior descending artery or its first diagonal branch, left circumflex artery or its first obtuse marginal branch and right coronary artery). The study also included control elderly males without a history of CAD or electrocardiographic signs of CAD. This group underwent percutaneous coronary intervention (PCI) or computer tomography coronary angiography to confirm normal coronary arteries. The controls were matched by age and geographical area to those in the case group. The exclusion criteria for case group or controls were as follows: 1) tumors; 2) thyroid diseases; 3) pulmonary hypertension; 4) heart failure; 5) secondary hypertension hypertrophic cardiomyopathy; 6) congenital heart disease; 7) atrial fibrillation; 8) infection; 9) autoimmune disease; or 10) severe hepatic dysfunction, renal dysfunction, or multiple organ failure based on medical history, physical examination, electrocardiography (ECG), echocardiography and biochemical measurements.
All of the subjects were evaluated using a detailed questionnaire that provided demographic data, coronary risk factors (e.g., smoking, diabetes mellitus, or hypertension), and family history of CAD. Smoking was classified as a consecutive or accumulative smoking history of more than 6 months, including current and ex-smokers. Diabetes mellitus was defined as fasting plasma glucose (FPG) ≥ 126 mg/dL (7.0 mmol/L), random plasma glucose ≥ 200 mg/dL (11.1 mmol/L), or 2-h plasma glucose ≥ 200 mg/dL (11.1 mmol/L). Hypertension was defined as a systolic blood pressure > 140 mmHg, diastolic blood pressure > 90 mmHg, or the use of any anti-hypertensive agent.
All of the data were collected by trained clinical research staff and were subsequently double-entered into computer databases. The study protocol was approved by the human rights committee of Chinese PLA General Hospital, and the study adhered to the Declaration of Helsinki and the Guidelines for Good Clinical Practice. Written informed consent for participation in the study was obtained from all participants.
Biochemical analysesBlood samples from a peripheral vein were obtained to examine biochemical parameters and to perform DNA extraction at the time of hospital admission. Blood samples were collected using vacutainer tubes and transferred to test tubes containing ethylenediamine tetra-acetic acid (EDTA). Genomic DNA was extracted using a standard phenol-chloroform method. Total cholesterol (TC), triglycerides, low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), fasting glucose and blood pressure were assessed using standard procedures.
Tag SNPs (tSNPs) selectionBased on CHB database (HapMap Data Rel27 PhaseⅡ+Ⅲ, population CHB) of the International HapMap Project (SNP browser v4.0, Applied Biosystems, Foster City, California, USA), we screened the PEDF gene locus spanning 22,108 bases from nucleotide position 1,607,009 to nucleotide position 1,629,117 (8 exons and 7 introns, located on human chromosome 17p13.3) as well as 5 kb of its 5′-flanking region and 1.5 kb of its 3′-flanking region. The linkage disequilibrium (LD) of SNPs was evaluated using Lewontin’s D′ and r2. The LD coefficient was calculated using the Haploview program, and haplotype blocks were defined with the same. Additionally, tSNPs were selected covering 100% of the common genetic variation (minor allele frequency, MAF > 0.1) within this locus with an r2 ≥ 0.8 based on Tagger analysis (http://www.broad.mit.edu/mpg/tagger).
DNA extraction and genotyping for SNPsBlood samples were collected and transferred to test tubes containing 100 μL 10% EDTA. The plasma was separated by centrifugation and the cellular portion was used for DNA extraction. A standard phenol-chloroform method (Rao et al. 2014) was used to extract DNA from peripheral leukocytes. The isolated genomic DNA was stored at −20℃ until genotyping.
Genotyping was performed by Shanghai Haotian Biotechnology Company utilizing ligase detection reaction (LDR) (Li et al. 2014). For each SNP, the alleles were distinguished using different fluorescent labels of allele-specific oligonucleotide probe pairs. Different SNPs were distinguished by different extended lengths at the 3′ end. The primers for both PCR and LDR reactions were all designed by Shanghai Haotian Biotechnology Company. Briefly, the PCR reactions were performed with 1 μL DNA sample (50 ng/μL), 5 μL 2 × GC buffer, 0.2 μL Mg2+ (25 mM), 1.2 μL dNTPs (2.5 mM), 0.07 μL HotStarTaq polymerase (5 U/μL), 1 μL multiple PCR primers (1 μM) and ddH2O in a total volume of 10 μL. The PCR cycling program was as follows: 95℃ for 2 min, followed by 1) 11 cycles of 94℃ for 20 s, 65℃ (decreased 0.5℃ per cycle) for 40 s, 72℃ for 90 s; plus 2) 24 cycles of 94℃ for 20 s, 59℃ for 30 s, and 72℃ for 90 s, with a final extension at 72℃ for 2 min. Then, 1 U shrimp alkaline phosphatase and 1 U Exonuclease I were added to the PCR products for purification. The LDR reactions were performed in a final volume of 10 μL containing 1 μL 10 × ligase reaction buffer, 0.5 μL Taq DNA ligase (200 U/μL), 0.5 μL of a 5′ ligase primer (1 μM) mixture, 0.5 μL 3′ ligase primer (1 μM) mixture, 3 μL purified PCR product, and 4.5 μL ddH2O. The LDR reactions were run for 38 cycles of 94℃ for 1 min and 56℃ for 4 min, and holding at 4℃. Then, 0.5 μL LDR product was sequenced with an ABI3130XL sequencer. Lastly, the raw data were analyzed using GeneMapper 4.1 (Applied Biosystems, USA). The primers for the seven target SNPs are shown in Table 1. The genotyping was carried out in a manner that was blind to group status. A random sample accounting for 5% (n = 40) of the total subjects was genotyped twice by different researchers for quality control, yielding a reproducibility of 100%.
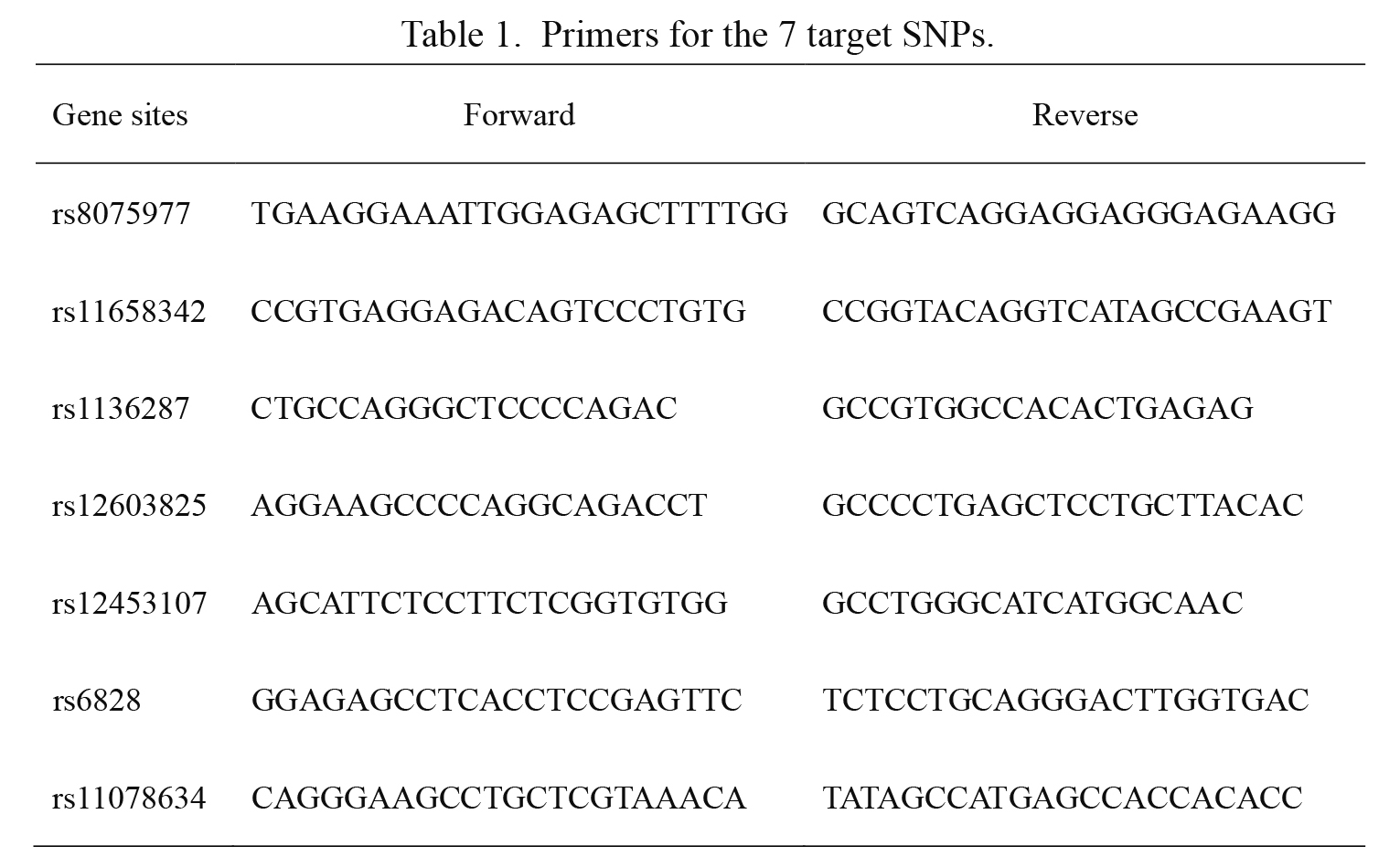
Primers for the 7 target SNPs.
Normally distributed continuous variables were expressed as the mean ± standard deviation (SD) and compared using a two-tailed Student’s t-test. Non-normally distributed data were presented as the median (P25-P75) and analyzed using nonparametric tests. Discrete numerical data are presented as frequency and percentage, and analyzed using the Chi-square test. The calculation of genotype and allele frequencies in CAD patients and healthy controls were determined by direct gene count method. The observed genotype frequencies were compared with the expected frequencies and tested for Hardy-Weinberg equilibrium (HWE) using the Chi-square test (Rao et al. 2014). The association between SNPs and the risk of CAD was investigated using the OR and 95% CI with logistic regression. Adjustments were made for risk factors, such as age, body mass index (BMI), hypertension, blood pressure etc. A Bonferroni correction was applied for multiple testing to reduce Type I errors. P < 0.007 was considered statistically significant with the adjusted significance level after Bonferroni correction. Other tests were two-sided, and P < 0.05 was considered statistically significant. All of the experimental results were analyzed using SPSS version 17.0.
An elderly Chinese Han male cohort including 416 CAD patients (mean age: 83.0 ± 8.9 years) and 528 controls (mean age: 81.3 ± 8.4 years) was enrolled in this case-control association study. The demographic data are summarized in Table 2. Patients with CAD were significantly older (P = 0.041), more likely to be hypertensive (P < 0.01), had greater prevalence of family disease history of CAD (P < 0.01), and exhibited higher systolic blood pressure (P = 0.043). Moreover, CAD patients exhibited significantly lower diastolic blood pressure (P = 0.048), TC levels (P < 0.01) and LDL-C levels (P < 0.01) compared to controls. There were no significant differences between CAD patients and controls with respect to BMI, diabetes mellitus, smoking history, HDL-C levels, triglycerides or fasting glucose.
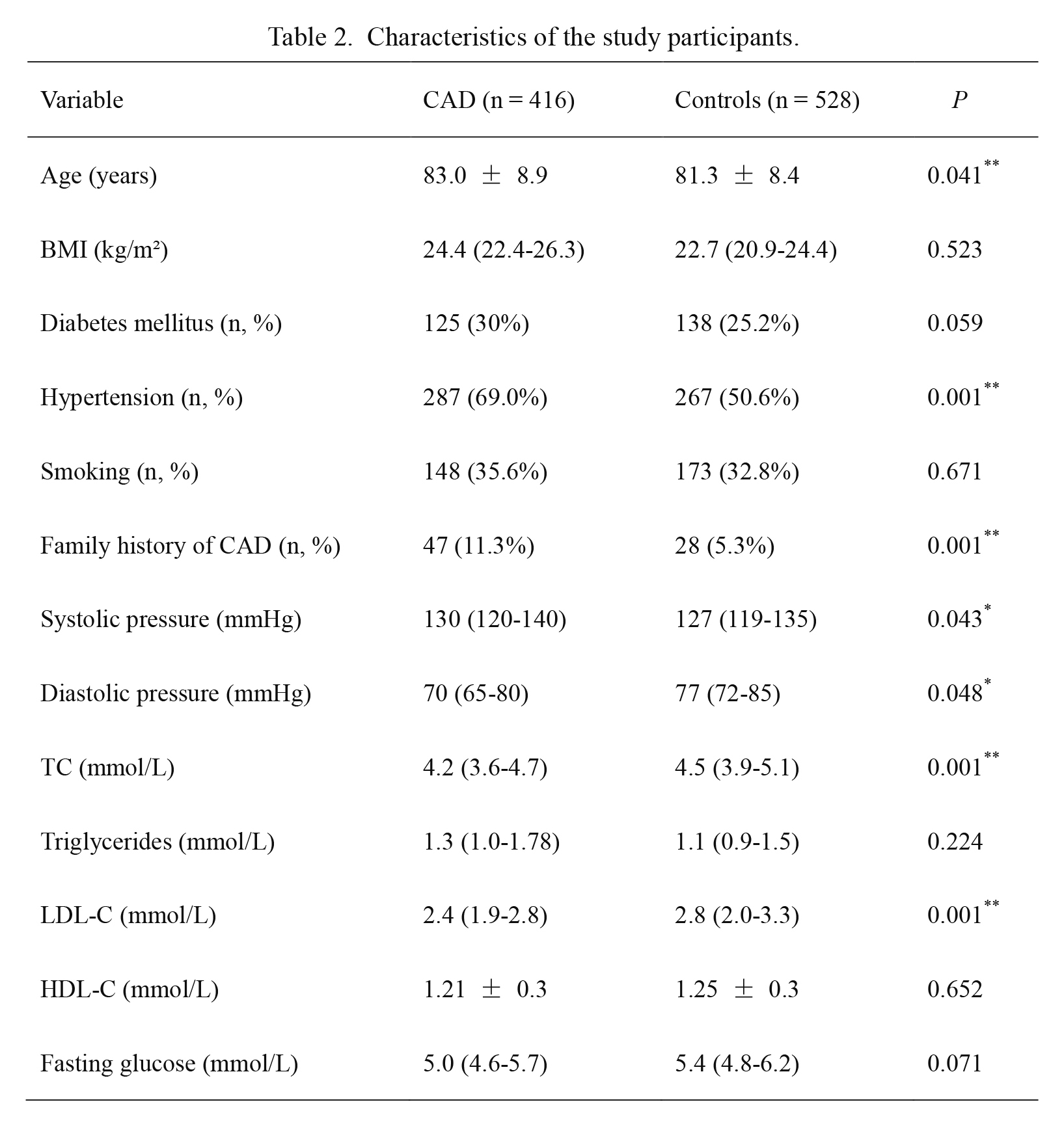
Characteristics of the study participants.
Bold values indicate statistical significance: P < 0.05 (*P < 0.05, **P < 0.01).
CAD, coronary artery disease; BMI, body mass index; TC, total cholesterol; LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol.
Seven PEDF tSNPs were selected, covering 100% of the common genetic variation (MAF > 0.1) within the PEDF locus with an r2 ≥ 0.8 based on Tagger program: rs8075977 in the 5′-flanking region, rs11658342 in the intron 2, rs1136287 in the exon 3, rs12603825 in the intron 3, rs12453107 in the intron 5, rs6828 in the exon 7 and rs11078634 in the 3′-flanking region (Table 3, Table 4). The overall genotyping success rates were > 99%. Genotype of each SNP is presented in Table 5.
HWE test revealed that genotype frequencies of each PEDF SNP did not significantly deviate from HWE expectations in total population (P > 0.05, Table 6), suggesting that the group populations were representative. We constructed a LD map for all the tested SNPs based on the genotyping data (Fig. 1A). To some degree, the LD map based on our genotyping data was essentially consistent with the LD map based on HapMap Data-CHB (Fig. 1B), suggesting the high accuracy of our genotyping results.

Characteristics of the PEDF (SERPINF1) tag SNPs.
The first column shows the tag SNP, which is a representative SNP that represents a group of SNPs in a block region with high linkage disequilibrium.
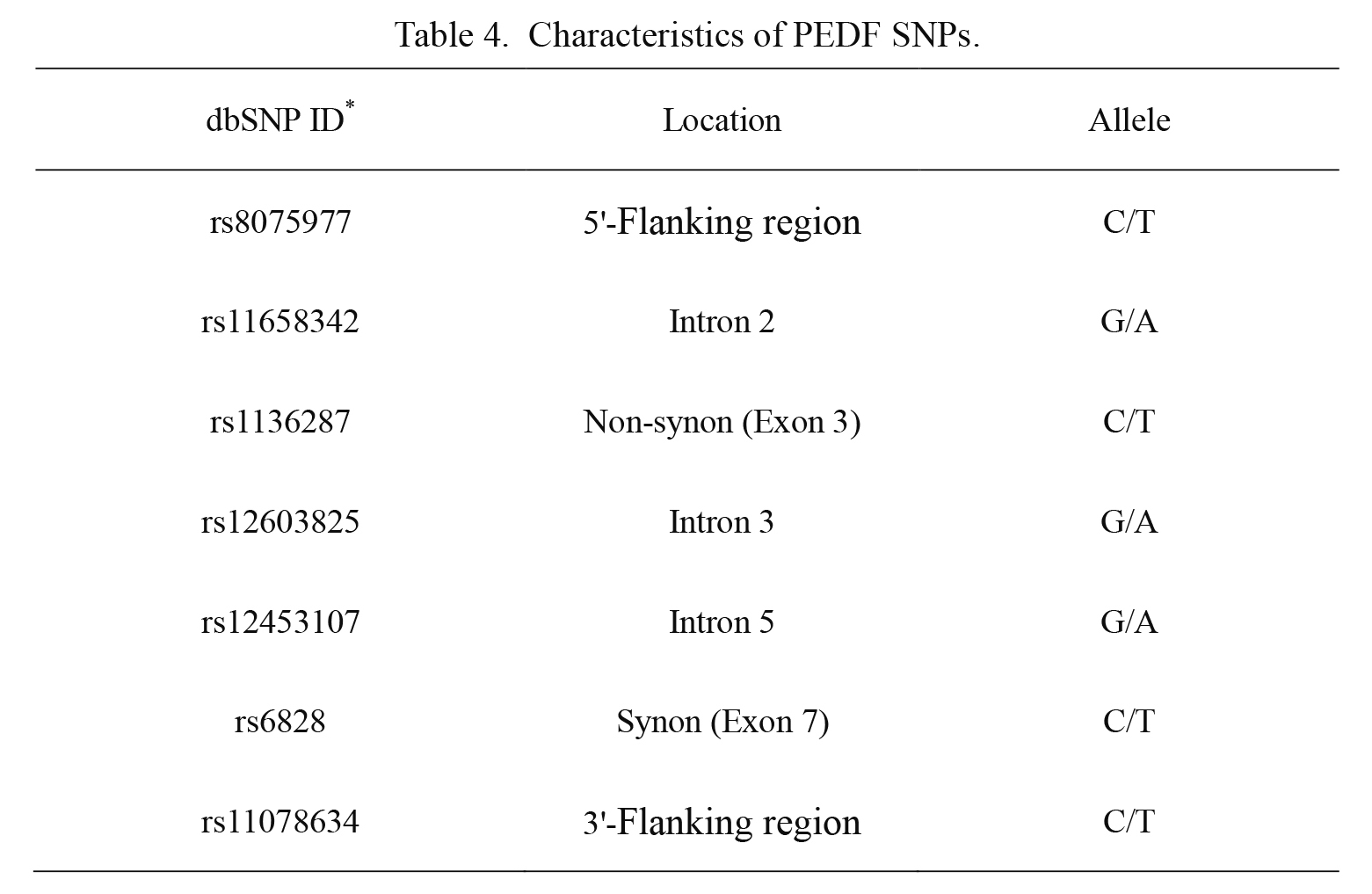
Characteristics of PEDF SNPs.
*dbSNP ID: the accession number of the polymorphic site on http://www.ncbi.nlm.nih.gov/SNP/.

The genotyping for PEDF SNPs.
Position 1 and Position 2 are peak positions of the two alleles of the SNP identified with Capillary Electrophoresis system. Genotypes of SNPs are based on the results of peak position.
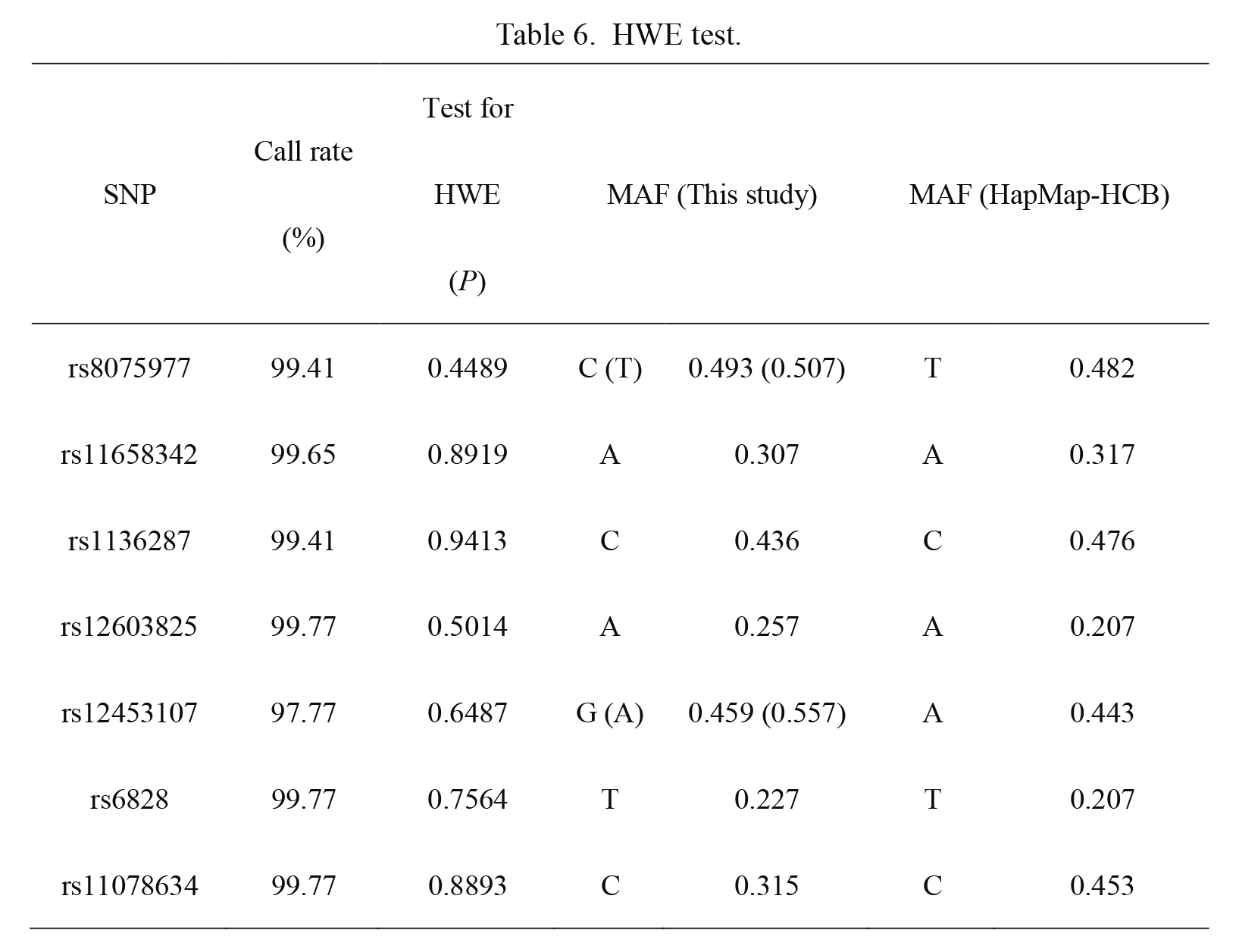
HWE test.
HWE, Hardy-Weinberg equilibrium; MAF, minor allele frequency.

Overview of LD structure among seven SNPs.
The PEDF gene on human chromosome 17p13.3 consists of 8 exons and 7 introns. The gene was screened spanning 22.11 kb from nucleotide 1,607,009 to nucleotide 1,629,117. The analyzed region included 5 kb of the 5′-flanking region and 1.5 kb of the 3′-flanking region. The locations of 7 tSNPs and 8 exons are indicated. The overall quality of genotyping data can be visually evaluated by LD map. A. The LD map was constructed based on the genotype data of all the subjects in the present study. B. The LD map was constructed based on HapMap Data-CHB. LD of SNPs was evaluated using Lewontin’s D′ and r2, and LD revealed the difference between the experimental and theoretical data.
LD, Linkage disequilibrium; SNPs, single nucleotide polymorphisms; PEDF, pigment epithelium-derived factor.
Information on the genotype and allele frequencies of the 7 SNPs in both groups is given in Table 7. Our data showed no significant differences in the genotype frequencies of the 6 PEDF SNPs (rs11658342, rs1136287, rs12603825, rs12453107, rs6828 and rs11078634) between the control and case groups. However, the frequency of rs8075977 T allele was 46.2% in patients with CAD, in contrast to 52.7% in the control group. The T allele frequency was significantly higher in the controls than in the case group (adjusted OR = 0.88, 95% CI: 0.80-0.96; P = 0.005).

The distribution of PEDF SNP allele and genotype frequencies in CAD patients and controls.
Bonferroni correction was applied for multiple testing to reduce Type I errors. P < 0.007 was considered statistically significant after adjustment for age, family history, hypertension, systolic pressure, diastolic pressure, TC, LDL and HDL. An adjusted OR and 95% CI were determined.
In this study, we genotyped seven PEDF SNPs to investigate any possible relationship between each SNP and CAD in elderly Chinese Han males. Seven tSNPs were identified, including rs8075977, rs11658342, rs1136287, rs12603825, rs12453107, rs6828 and rs11078634. Among them, only one SNP, rs8075977 (C/T) located in the 5′-flanking region, showed the significant effect on the susceptibility to CAD in elderly Chinese Han males. Moreover, the T allele at this common variation site may be a protective factor for CAD.
Given the global burden of cardiovascular disease (CVD), there has been an increasing interest in the pathogenesis, treatment and prevention of this condition, as well as CAD and atherosclerosis. Studies of the etiology and pathogenesis of CAD have advanced to the level of genomic analysis with improvements in molecular genetics techniques. Gene polymorphisms are known to modulate the functions of certain gene products, and various epidemiological studies in families and twins have revealed the crucial role of genetic factors in CAD. In addition, several gene variants involved in pathogenesis of atherosclerosis, including inflammation, oxidative stress and thrombogenesis, can contribute to CVD risk (Wu et al. 2014). Therefore, much of previous work has concentrated on the relationships of certain genes and polymorphisms with CAD. The MTHFR rs1801133 (C/T) polymorphism was found to be associated with CAD development (Chen et al. 2014). It was reported that the GG genotype of rs12671368 and the AA genotype of rs2192932 in the MLL5 gene could be protective genetic markers of CAD (Yuan et al. 2014). In addition, there is evidence showing that the HNF1A rs7310409 (G/A) functional polymorphism may contribute to the risk of left main CAD (Liu et al. 2014d). This study also stated that inflammation was responsible for the initiation and progression of CAD and atherosclerosis. C-reactive protein (CRP) is one of many important inflammatory factors involved in CAD (Lagrand et al. 1999), and investigations have reported a significant association between HNF1A SNPs and CRP levels (Reiner et al. 2009). Similar to the majority of these studies, we also analyzed polymorphisms in the present study.
PEDF was originally detected in the growth medium of human fetal retinal pigment epithelial cells (Tombran-Tink and Johnson 1989). This protein has been found to exert anti-inflammatory, anti-oxidant, anti-thrombotic and anti-angiogenic effects. Accordingly, PEDF may be a potential target molecule for preventing the progression of atherosclerosis. In addition, PEDF is involved in CAD and is attracting more attention. Apart from its properties as a multifunctional glycoprotein, PEDF is a reasonable candidate gene for particular diseases. Indeed, a role of PEDF genetic polymorphisms has been observed in numerous diseases, such as AMD, myopic choroidal neovascularization, insulin resistance and type 2 diabetes. On the grounds of common disease/common variant (CDCV) model, common genetic variants of small to moderate effect underlie the risk of common disorders or diseases (Uher 2009). Furthermore, both abnormal angiogenesis in a multitude of ophthalmic diseases as mentioned above and obesity-related insulin resistance are related to atherosclerosis. Therefore, the clear association is putatively important between CAD and polymorphisms of PEDF with anti-inflammatory, anti-oxidant, anti-thrombotic, and anti-atherosclerotic properties.
We performed a case-control cohort study, and the demographic data showed that CAD patients were significantly older (P = 0.041) than the control group and were more likely to be hypertensive (P < 0.01), have a family history of CAD (P < 0.01) and have higher systolic pressure (P = 0.043). Nevertheless, diastolic pressure, TC and LDL-C levels of CAD patients were significantly lower than in the control group (P = 0.048, P < 0.01 and P < 0.01, respectively). The patients were selected from a population of elderly males, who therefore may be diagnosed with hypertension, hyperlipidemia or CAD many years ago. It is possible that the data were affected to some degree by the medications that these individuals took on a daily basis. Next, a tag approach was used to cover 100% of common variations in the PEDF gene (MAF > 0.1). We ultimately selected 7 tSNPs (rs8075977, rs11658342, rs1136287, rs12603825, rs12453107, rs6828 and rs11078634) using the HapMap Data-CHB and Tagger program. In addition, genotyping for the 7 SNPs was successfully performed (Table 4). Our data of 7 SNPs included PEDF rs1136287 (C/T) and rs11658342 (G/A), two polymorphisms which were also selected as PEDF tSNPs in the study of exudative AMD (Qu et al. 2011). This study also showed that heterozygosity for rs1136287 (C/T) was protective against exudative AMD (additive model, OR = 0.59, 95% CI: 0.36-0.95; P = 0.03) (Qu et al. 2011). Moreover, it has been demonstrated that rs12603825 (G/A) and rs6828 (C/T) are tSNPs for PEDF and only rs12603825 (G/A) is significantly associated with MRI-derived total adipose tissue mass (P = 0.0094) and fasting leptin concentrations (P = 0.0035); this SNP rs12603825 (G/A) was also found to be nominally associated with body fat percentage as determined by bioelectrical impedance (P = 0.0182) and clamp-derived insulin sensitivity (P = 0.0251) (Bohm et al. 2012). Although rs1136287 (C/T), rs11658342 (G/A), rs12603825 (G/A) and rs6828 (C/T) were also selected as PEDF tSNPs in the present study, there was no correlation between each of these four SNPs and CAD. Instead, among the 7 SNPs, only the frequency of rs8075977 T allele was significantly different between CAD and control subjects (adjusted OR = 0.88, 95% CI: 0.80-0.96; P = 0.005) after Bonferroni correction.
Our results showed that the frequency of SNP rs8075977 T allele was 0.462 in CAD and 0.527 in controls (Table 7), suggesting that T residue can be considered as a common sequence. Apparently, this T allele frequency was significantly higher in controls than in CAD patients. Maybe the reasons are as follows: (1) rs8075977 is really correlated with CAD; (2) there is a strong level of LD between rs8075977 and a polymorphic loci which functions as the real protection against CAD. PEDF SNP rs8075977 is on chromosome 17. Human chromosome 17 is implicated in a wide range of human genetic diseases including early-onset breast cancer (BRCA1), neurofibromatosis (NF1) and the DNA damage response (TP53 encoding the p53 protein) (Zody et al. 2006). On this potential genetic foundation, together with the evidence of a potential protective effect of PEDF against CVD or atherosclerosis (Rychli et al. 2009; Wang et al. 2013; Liu et al. 2014d) and the effect of SNP on gene expression (Samnegard et al. 2005), we speculate that the observed correlation of this SNP rs8075977 (C/T) with CAD may be due to an influence on PEDF expression. As shown in Table 4, 7 PEDF SNPs have different location: flanking sequence, intron and exon. Genetic polymorphisms in coding sequence can result in amino acid changes and subsequently affect the activity or expression levels of a target protein (Moreno et al. 2004; Lin et al. 2008). PEDF SNP rs1136287 (C/T), one of our genotyping results, has been identified a functional amino acid change. This SNP is a methionine to threonine polymorphism (Met72Thr polymorphism) in exon 3 of the PEDF gene that causes the formation of BsstSI restriction site, and the PEDF Met72Thr T allele may be a risk factor for wet AMD (Koenekoop et al. 1999; Lin et al. 2008). By contrast, our data showed that there was no association between SNP rs1136287 (Met72Thr) and CAD. The altered PEDF activity or property via changed amino acid affected by the polymorphism could be a conceivable explanation. Flanking sequence is known to contain enhancer, terminater and promoter. Indeed, SNPs in promoter region maybe a mechanistic explanation for some diseases. The different location may be one reason why only rs8075977 in the 5′-flanking region was associated with CAD in Chinese Han elderly men compared with the other 6 SNPs.
SNP rs8075977 (17: 1660801, C/T, intergenic SNP) is located in the 5'-flanking region of the PEDF gene. The gene SNP annotation results showed that rs8075977, 4.4 kb upstream of the transcription start site of PEDF gene, is not a promoter SNP. However, cases of long-range interactions involved in activation and repression of transcription have been obtained in a variety of organisms, suggesting that gene regulation through direct physical association with regulatory elements and/or other genes is a common and conserved phenomenon (Miele and Dekker 2008). Therefore, we supposed that rs8075977 may affect PEDF gene transcription instead of the function or stability of PEDF. Given the recent functional researches on loci or diseases (Ohnishi et al. 2007; Li et al. 2016), SNP rs8075977 may be on the transcription factor binding sites and presumably alters PEDF expression by binding a certain transcription factor. Thus we analyzed rs8075977 using publically available software Haploreg (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php). Significantly, the prediction analysis showed that rs8075977 is likely to be on the binding site of transcription factor IIA (TFIIA) and to be related to the expression of TLC domain-containing 2 (TLCD2) (Fehrmann et al. 2011) and SET and MYND domain containing 4 (SMYD4) (Westra et al. 2013). TFIIA, the general transcription factor, can activate accurate transcription initiation through stabilization of TATA box-bound TBP (TATA-binding protein) (Suzuki et al. 2015). In addition, TFIIA can regulate responses to transcriptional activators and be recruited by transcriptional activators to specific target promoters (Kim et al. 2008). Interestingly, TFIIA was reported to function in the cellular mechanism of defense against ROS, suggesting its role in the response to oxidative stress (Kraemer et al. 2006), which is in agreement with the anti-oxidant property of PEDF (Rychli et al. 2009). Although rs8075977 is present within the binding site of TFIIA, it remains unknown whether this SNP disrupts the consensus binding-sequence for TFIIA. TLCD2 may be involved in ceramide synthesis, lipid metabolism and cardiac hypertrophy (Winter and Ponting 2002; Pewzner-Jung et al. 2006; Harper et al. 2013). Furthermore, TLCD proteins are thought to function as sphingolipid sensors involving in lung inflammation (Winter and Ponting 2002; Uhlig and Gulbins 2008; Nixon 2009) and data suggested that TLCD2 could be a candidate negative regulator of eosinophil recruitment responses to allergen (Kelada et al. 2014). Despite no direct evidence of the correlation between PEDF and TLCD2, they are common in cardiovascular diseases and inflammation to some degree. SMYD, a methyltransferase, is involved in epigenetic transcription, cell proliferation and the development of cancer, heart and skeletal muscle (Du et al. 2014; Calpena et al. 2015). It is a putative intervention target for carcinomas and cardiovascular diseases (Spellmon et al. 2015). A drosophila SMYD4 homologue is manifested to be a muscle-specific transcriptional modulator (Thompson and Travers 2008). Additionally, SMYD4 was identified as a potential tumor suppressor in breast cancer (Hu et al. 2009), similar to PEDF that is a potential therapeutic target with multiple anti-cancer activities (Ek et al. 2006). Although the functions of TLCD2 and SMYD4 need to be further clarified, these researches above seem to associate SNP rs8075977 with PEDF expression more directly and closely. In fact, a variety of very complicated factors can affect gene expression or regulation, including transcription factors. To exactly demonstrate the effect of SNPs on transcription factor binding sites and transcription rate, more functional studies, luciferase reporter assay and real-time quantitative PCR for example, are warranted.
The present study had several strengths and limitations. Genotyping errors are the most common and severe problem in SNP studies (Pompanon et al. 2005). One strength of the present research was that the genotyping and analysis were performed under rigid quality controls, such as the MAF test, HWE test, LD test, and repeatability test; moreover, there was a high-rate of successful genotyping. Another strength lies in the subjects, who were all elderly Chinese Han males (age ≥ 60 years). First, this homogeneity helped to reduce false positives and weakened any effects of differences in genetic background; these complications cannot be eliminated by statistical adjustments. Second, males are more susceptible to atherosclerosis and CAD than females. Last but not least, the population as a whole is aging, resulting in an increase in global cardiovascular deaths over the past two decades (Roth et al. 2015). Elderly patients with CVD should be given more attention than ever before. One limitation of the study is that the number of enrolled participants was limited. Although we were able to limit false positives, the sample size was relatively small, as was the population from which the participants were selected. Moreover, it is possible that selection bias occurred. The second limitation of the study was that it had a single focus. Association studies between PEDF SNPs and PEDF serum levels, CAD diagnosis, CAD severity, and disease prognosis should be carried out. The results of these studies may be more conducive to understanding the genetic characteristics of CAD.
In conclusion, the PEDF SNP rs8075977 may be associated with CAD susceptibility in elderly Chinese Han males. Moreover, the T allele of rs8075977 may be protective for CAD. The data increase our understanding of how genetic variants influence CAD and support a role for PEDF in the pathogenesis of CAD. Larger studies in more ethnically diverse populations are needed to confirm these findings.
This work was supported by Logistics scientific research project (No. 15BJZ37) and the National Natural Science Foundation of China (Grant nos. 81470504).
The authors declare no conflict of interest.