2018 Volume 244 Issue 2 Pages 113-117
2018 Volume 244 Issue 2 Pages 113-117
Host-derived factors alter gut microenvironment, and changes in gut microbiota also affect biological functions of host. Alterations of gut microbiota have been reported in a wide variety of diseases, but the whole picture of alterations in pancreatic diseases remains to be clarified. In particular, the gut microbiota may be affected by malnutrition or impaired exocrine pancreas function that is associated with pancreatic diseases. We here conducted comprehensive analysis of gut microbiota in patients with type 1 autoimmune pancreatitis (AIP), a pancreatic manifestation of the systemic IgG4-related disease, and chronic pancreatitis (CP). The two diseases were selected, because altered immune reactions in AIP and/or long-standing malnutrition in CP may influence the gut microbiota. Fecal samples were obtained from 12 patients with AIP before the steroid therapy and 8 patients with CP. Metagenome DNA was extracted, and microbiota was analyzed by next generation sequencing. Gut microbiota profiles were different between patients with AIP and those with CP; namely, the proportions of Bacteroides, Streptococcus and Clostridium species were higher in patients with CP. The reasons for the increased proportion of these bacterial species remain unknown, but may reflect malabsorption and/or decreased pancreatic enzymes, both of which are associated with CP. Incidentally, the identified Streptococcus species are oral cavity inhabitants and also known as pathogens for endocarditis. Despite the small sample size, this study has shown the differences in gut microbiota profiles between AIP and CP. Comprehensive analysis of the gut microbiota may be useful for the differential diagnosis of pancreatic diseases.
Interactions between the host and gut microbiota alter the immune system and other biological functions, contributing to the pathogenesis of several diseases. For example, imbalance of gut microbiota, referred to as dysbiosis, is associated with multiple sclerosis, a chronic demyelinating disease triggered by an autoimmune mechanism (Yadav et al. 2017). Subgingival dysbiosis has been reported in patients with systemic lupus erythematosus, another autoimmune disease (Correa et al. 2017). In the field of gastroenterology, it has been recognized that alterations in gut microbiota have substantial roles in inflammatory bowel diseases (Alipour et al. 2016). Dysbiosis in gut microbiota causes altered bile acid composition, leading to the loss of the anti-inflammatory effects of bile acid (Duboc et al. 2013). Alterations of the microbiome have certain causative roles and normalization of the microbiome might have therapeutic applications. Fecal microbiota transplantation might be useful for the treatment of inflammatory bowel diseases as well as recurrent Clostridium difficile infection (Kelly et al. 2016; Reinisch 2017).
Alterations of the gut microbiota could serve as a disease marker (Wen et al. 2017). A recent study showed decreased bacterial diversity in patients with Alzheimer’s disease, accompanied by a reduction of Bifidobacterium (Vogt et al. 2017). A similar phenomenon has been reported in patients with ankylosing spondylitis, with increases in Prevotella melaninogenica, Prevotella copri, and Prevotella sp. Detection of fecal bacteria subsets such as Fusobacterium nucleatum, Bacteroides clarus and Roseburia intestinalis successfully identified patients with colorectal cancer, suggesting a potential application of microbiota profiles as a novel diagnostic biomarker (Liang et al. 2017).
Pancreatic diseases such as pancreatic cancer, chronic pancreatitis (CP) and autoimmune pancreatitis (AIP) are often accompanied by pancreatic exocrine insufficiency (Hart et al. 2015; Ramsey et al. 2017; Vujasinovic et al. 2017), which could affect the gut microenvironment and microbiota. AIP is a rare type of pancreatitis with a hypothesized autoimmune mechanism (Hart et al. 2015; Okazaki et al. 2017). It has two distinct phenotypes, termed type 1 and type 2. Type 1 AIP is currently regarded as a pancreatic manifestation of the systemic immunoglobulin G4-related disease (Hart et al. 2015). A recent systematic review has revealed a role of gut microbiota in pancreatic cancer, but the information on pancreatitis was limited (Memba et al. 2017). Impaired acinar cell function resulted in bacterial overgrowth and dysbiosis in a mouse model (Ahuja et al. 2017). It is reasonable to assume that altered immune reactions in AIP and long-standing malnutrition in CP modify the dysbiosis. In addition, it would be of interest to see whether gut microbiota profiles are useful to diagnose early CP, distinguish AIP from CP, and predict disease relapses in AIP (Hamada et al. 2015; Masamune et al. 2017a, b). We here report comprehensive analysis of gut microbiota in patients with AIP and those with CP.
Twelve patients with type 1 AIP before the steroid treatment and 8 patients with CP were enrolled. This study was approved by the Ethics Committee of Tohoku University Graduate School of Medicine (article#: 2016-1-316).
DNA extractionAfter written informed consent was obtained, small fecal samples were collected in a tube for the fecal occult blood test, and stored at –80°C until use. Fecal DNA was extracted using PowerFecal DNA isolation kit (MO BIO Laboratories, Carlsbad, CA) according to the manufacturer’s protocol. The extracted DNA samples were dissolved in 100 μl of TE buffer, resulting in a final DNA concentration of less than 10 ng/μl.
Real-time PCR for bacterial 16S rRNA geneThe amount of bacterial 16S ribosomal RNA (rRNA) gene in each 2 μl DNA sample was quantified by real-time PCR using the following primer set and probe (Ritalahti et al. 2006). The sequences were 5′-ATGGYTGTCGTCAGCT-3' (Bac1055YF) for the forward primer, 5'-ACGGGCGGTGTGTAC-3′ for the reverse primer (Bac1392R), and 5′-FAM-CAACGAGCGCAACCC-TAMRA-3′ (Bac1115Probe) for the probe. Due to the low concentration of DNA sample, copy number per volume of DNA solution derived from same amount of stool sample was calculated.
Next-generation sequencing for microbiome analysisThe V4-V5 region of the bacterial 16S rRNA gene was amplified from 2 μl fecal DNA samples. PCR products were subjected to 250 bp of pair-end analysis using MiSeq (Illumina, San Diego, CA). After trimming low-quality sequence data, similar (97% or more similarity) sequence data were clustered as an operation taxonomic unit (OTU). Each OTU was subjected to database analysis using Greengene database and Living Tree (DeSantis et al. 2006; Caporaso et al. 2010; Munoz et al. 2011). Each OTU’s taxonomic classification was confirmed and read counts in each sample were compared. Proportions of each bacterial species in total read counts were then determined.
Statistical analysisStatistical analysis was performed using JMP (SAS Institute, Cary, NC). The differences between two groups were analyzed using the t-test or Fisher’s exact test. A P value of < 0.05 was regarded as statistically significant.
Patients’ age and the ratio of males did not differ between AIP and CP (Table 1). Proportions of smokers and drinkers tended to be higher in CP patients, but it was not statistically significant (Table 1). Extracted DNA samples were subjected to quantification of 16S rRNA gene. Even from DNA samples of less than 10 ng/μl of DNA, sufficient 16S rRNA gene could be amplified. There was no difference in the 16S rRNA gene copy number between the patients with AIP and CP (Fig. 1).
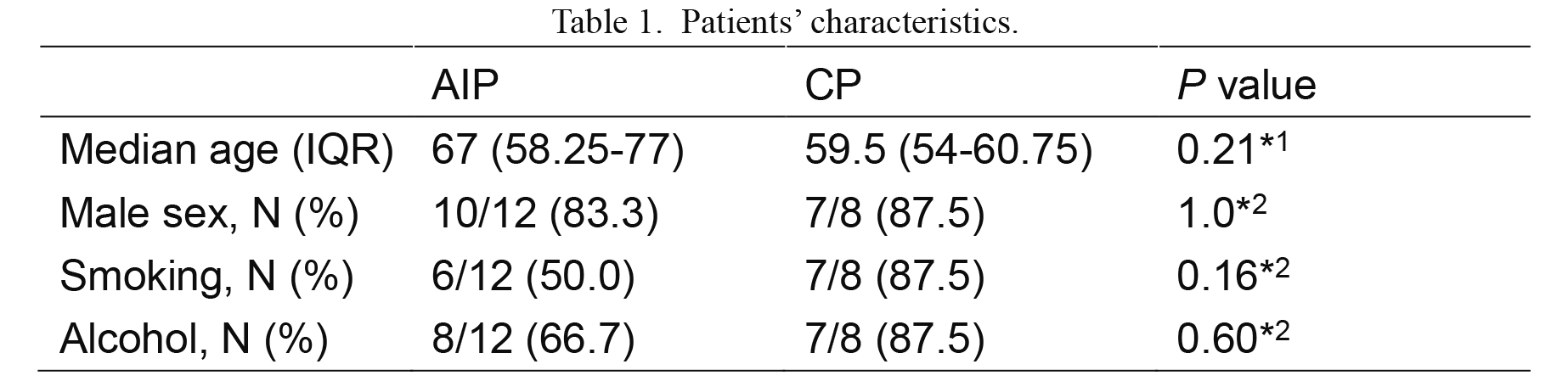
Patients’ characteristics.
*1t-test, *2Fisher’s exact test.
IQR, interquartile range.

Quantification of bacterial 16S rRNA gene in fecal samples.
Copy number of bacterial 16S rRNA gene in fecal samples from patients with AIP before the steroid treatment (N = 12) and CP (N = 8). n.s., not significant.
To identify difference in gut microbiota between AIP and CP, we compared microbiome profiles in these patients. At the phylum level, there was no difference between the AIP and CP patients (data not shown). However, several species were detected more frequently in CP patients. Bacteroides ovatus, Streptococcus australis, Streptococcus gordonii, Clostridium lactatifermentans, Clostridium lavalense were more frequent in CP patients compared to AIP patients (Fig. 2). Oki et al. (2016) reported that the most predominant bacterial families in fecal microbiota were Bacteroidaceae, Lachnospiraceae, Ruminococcaceae, and Prevotellaceae in healthy Japanese adults. Bacteroides ovatus stimulated normal colonic mucosa for cytokine production, suggesting possible involvement in inflammation (Sundin et al. 2015).

Identification of increased bacterial species in CP.
Proportions of bacterial species in gut microbiota were determined in patients with AIP before the steroid treatment (N = 12) and CP (N = 8). A, Bacteroides ovatus; B, Streptococcus australis; C, Streptococcus gordonii; D, Clostridium lactatifermentans; E, Clostridium lavalense. *P < 0.05, **P < 0.01.
This study showed differences in gut microbiota between patients with CP and those with AIP. To our knowledge, this is the first study to assess the gut microbiota in patients with AIP. The stool sampling method for fecal blood testing could provide a large metagenome sample sufficient for the microbiome analysis. Several bacterial species were more abundant in the gut microbiota of CP patients compared to those of AIP patients. Among them, Streptococcus australis and Streptococcus gordonii are generally present in the oral cavity, sometimes causing bacterial endocarditis (Warburton et al. 2013; Dadon et al. 2017). A greater abundance of Streptococcus has also been found in patients with intestinal diseases in a meta-analysis (Mancabelli et al. 2017), suggesting a possible contribution of these bacterial species during the dysbiosis of CP patients. The reasons for the increased proportion of these bacterial species remain unknown, but might include malabsorption or decreased pancreatic enzymes associated with CP. In contrast, the proportion of butylate-producing specie such as Clostridium lavalense, which has been reported to be reduced in Crohn’s disease (Takahashi et al. 2016), was increased in CP patients. Jandhyala et al. (2017) reported that the Firmicutes: Bacteroidetes ratio was increased in CP patients. Whether these findings are adaptive phenomena or related to the pathogenesis of CP needs further validation.
We here focused on AIP and CP, because international consensus on the definition of AIP as a subtype of CP has not been established. The Japanese diagnostic criteria for CP excluded AIP from CP because AIP is reversible (Shimosegawa et al. 2010), whereas AIP was defined as a subtype of CP in other classifications (Etemad and Whitcomb 2001; Schneider et al. 2007). Microbiota profiles might be useful for the differential diagnosis of pancreatic diseases and patient subgroups in the future.
The current study has several limitations due to the small number of patients enrolled. In addition, functional roles of identified bacterial species and relevance to pathogenesis in AIP and CP remain unclear. Further validation and assessment of the efficacy are necessary to differentiate AIP and CP based on the microbiome profile. A recent report suggested that the nutritional status also affects the gut microbiota in mice. Administration of pancreatic digestive enzymes to mice altered the composition of the gut microbiota (Nishiyama et al. 2018). Pancrelipase treatment increased Akkermansia muciniphilaI and Lactobacillus reuteri, which are beneficial bacteria. Similar changes were not detected in the current study, which might reflect malabsorption or because dysbiosis commonly exists in both AIP and CP. To address this issue, a comparison between AIP patients before and after steroid therapy, or CP patients with or without pancreatic enzyme supplementation might be important. Importantly, a precise assessment method for exocrine pancreatic function is not routinely available in Japan.
In conclusion, we reported the comprehensive analysis of gut microbiota in patients with AIP and CP using small stool samples. Such analysis for diagnosis, clarification of the contribution of the microbiome to the pathogenesis and use as a possible tool for fine-tuning of the treatment should be assessed in the future.
This work was supported in part by JSPS KAKENHI (16K15421, 17K09452, 15H04804), and the Smoking Research Foundation (to Masamune, A.).
The authors declare no conflict of interest.