Abstract
The Tohoku Medical Megabank biobank (TMM biobank) is the first major population-based biobank established in Japan. The TMM biobank was established based on two population cohorts and is a reconstruction program from the Great East Japan Earthquake and Tsunami of 2011. The biobank stores more than 3.4 million tubes of biospecimens and associated health and analytic data obtained from approximately 150,000 TMM cohort participants between May 2013 and December 2018, and the TMM biobank currently shares high-quality specimens and data. Various biospecimens, including peripheral and cord blood mononuclear cells, buffy coat, plasma, serum, urine, breast milk and saliva have been collected in the TMM biobank. To minimize human error and maintain the quality of data and specimens, we have been utilizing laboratory information management system into various biobank procedures from registration to storage with various automation systems, such as liquid dispensing, DNA extraction and their storage. The biobank procedures for the quality management system (ISO 9001:2015) and information security management system (ISO 27001:2013) are certified by the International Organization for Standardization. The quality of our biobank samples fulfills the pre-analytical requirements for researchers conducting next-generation whole genome sequencing, DNA array analyses, proteomics, metabolomics, etc. We established analytical centers to conduct standard genomic and multiomic analyses in-house and share the generated data. Additionally, we generate thousands of Epstein-Barr virus (EBV)-transformed lymphoblastoid cell lines and proliferating T cells for functional studies. The TMM biobank serves as an indispensable infrastructure for academic, clinical and industrial research to actualize next-generation medicine in Japan.
Introduction
The Great East Japan Earthquake and succeeding tsunami on March 11, 2011 damaged Pacific Coast provinces of East Japan. In particular, three prefectures, Iwate, Miyagi and Fukushima, suffered severely. Reports indicate that 15,897 people were lost and 2,533 people are still missing in these prefectures (National Police Agency 2019). In addition, medical infrastructures in these areas were devastated. To address the medical problems in the damaged areas and to establish an advanced medical system in the tsunami-affected areas, we proposed the Tohoku Medical Megabank (TMM) Project in 2011.
The TMM Project was funded and started in February 2012. An aim of the project was to establish two large population cohorts and a biobank based on these cohorts. Together, the cohorts and biobank will establish a solid foundation for future medicine, including personalized healthcare and genomic medicine (Kuriyama et al. 2016; Yasuda et al. 2019). For the cohort studies, we established a TMM community cohort (TMM CommCohort) study and a TMM birth-and-three-generation cohort (TMM BirThree cohort) study. Recruitment to the CommCohort study started in May 2013. This cohort study was conducted in both the Miyagi and Iwate prefectures, with an aim to recruit 50,000 and 30,000 participants, respectively, from each prefecture (Kuriyama et al. 2016). During the first-phase recruitment period, until 2017, more than 87,000 participants were recruited to the CommCohort study. The BirThree cohort started preliminary recruitment in July 2013, which accelerated in early 2014. The BirThree cohort was conducted mainly in Miyagi Prefecture, and during the first-phase recruitment period (until 2017), more than 73,000 individuals gave consent for inclusion in the cohort study (Kuriyama et al. 2016). In total, we collected specimens from approximately 4.1% of the entire population of the Miyagi and Iwate prefectures.
The TMM biobank has been collecting various biospecimens, including peripheral and cord blood mononuclear cells, buffy coat, plasma, serum, urine, breast milk, and saliva, from its participants. The quality of our biobank samples fulfills the preanalytical requirements for researchers conducting next-generation whole-genome sequencing, DNA array analysis, proteomics, metabolomics, etc. Notably, we established an analytical center within the TMM Project and conducted standard genomic as well as multiomics analyses in-house. We refer to the system that combines the biobank and analytical center as an integrated biobank. We share the generated data widely, as the basic cohort data and genome-omics big data are managed under one regulatory system, the TMM database (Database Tohoku Medical Megabank, or dbTMM). In addition, our TMM biobank generates and stores thousands of Epstein-Barr virus (EBV)-transformed lymphoblastoid cell lines and proliferating T cells for further functional studies.
As the biobank that organizes advanced large-scale population-based studies, the quality of the biospecimens and analyzed data, as well as the integrity of the associated data, are critical. Here, we describe the design and systems of the TMM biobank that guarantee quality and integrity and the present status of biobank management. The TMM biobank is the first large-scale population-based biobank established in Japan. It is therefore valuable to introduce aims, concepts and progress of the TMM biobank systems, such that how we collect, store, select and share biospecimens for various advanced genome- and multi-omics-oriented medial, informatic and biological studies. We believe that the TMM biobank will serve as an indispensable infrastructure for academic, clinical, and industrial research that actualizes next-generation medicine in Japan.
Materials and Methods
Transportation, central processing and storage
Biospecimens collected in Miyagi Prefecture were transported to the TMM biobank on the day of collection, while those collected in Iwate Prefecture were received the following morning. During transportation, all biospecimens were stored in boxes with temperature data loggers under controlled temperature conditions (4°C or 20°C). These conditions have been validated to be suitable for omics studies (Koshiba et al. 2016; Saigusa et al. 2016). Samples were immediately deidentified on arrival, as described (Takai-Igarashi et al. 2017).
Information management system
We used a partially customized LabVantage laboratory information management system (LIMS) (ver. 6 and ver. 8; LabVantage Solutions, Somerset, NJ, USA).
Sample processing
A 700 μL volume of each liquid sample, including serum, plasma, urine and milk, was automatically aliquoted into 1-mL Matrix 2D barcoded storage tubes (BC30661; identical to Thermo Fisher Scientific 3741JP but with a custom barcode for the latch racks, Thermo Fisher Scientific, Waltham, MA, USA) by Freedom EVO 200 and 150 robots (Tecan Group, Männedorf, Switzerland) (Mathay et al. 2012). In addition, buffy coat samples from blood and cord blood were aliquoted into two tubes (490 and 210 μL) with the robots. We manually processed samples with 1) a small volume or 2) blood clotting. All 2D tubes were placed in a –80°C freezer for more than one night and were then moved into a Brooks BioStore automated sample storage system (Brooks, Chelmsford, MA, USA) at –80°C.
Peripheral blood mononuclear cells (PBMCs) were isolated using BD Vacutainer CPT tubes according to the manufacturer’s recommendations. The collected PBMCs were then resuspended in 1 mL of CELLBANKER 1 plus (Nihon Zenyaku Kogyo, Koriyama, Japan). Cell numbers were determined using a fully automated blood cell counter (Celltac F, Nihon Kohden, Tokyo, Japan). PBMCs were dispensed into two sterile 1-mL Matrix 2D tubes in 500-µL aliquots, immediately frozen in prechilled Bicell (Nihon Freezer, Tokyo, Japan) at –80°C and kept overnight before being stored in –180°C tanks of vapor-phase liquid nitrogen storage systems (Chart-MVE Biomedical, Canton, GA, USA and Taiyo Nippon Sanso, Tokyo, Japan).
Establishment of EBV-transformed lymphoblastoid cell lines and activated T cells
CD19-positive cells were isolated from one tube of cryopreserved PBMCs described above using an EasySep Human CD19 Positive Selection kit (STEMCELL Technologies, Vancouver, Canada) according to the manufacturer’s recommendations. The cells were infected with EBV-containing culture supernatant from EBV-producing marmoset cells (B95-8) (Neitzel 1986). Infected CD19-positive cells were resuspended in 100 µL of complete RPMI 1640 medium containing 20% heat-inactivated fetal bovine serum (FBS, Sigma-Aldrich, St. Louis, MO, USA), 100 U/mL penicillin and 100 µg/mL streptomycin (Nacalai Tesque, Kyoto, Japan), 2 mM GlutaMAX I, and MEM containing nonessential amino acids (Thermo Fisher Scientific) and incubated in a 96-well plate at 37°C in 5% CO2. By one week after EBV infection, clusters of cells were visible by light microscopy. As cells developed into EBV-LCLs (EBV-lymphoblastoid cell lines), the microscopic clusters enlarged, and clumps were visible macroscopically. Subsequently, the culture was expanded by increasing the culture volume by 2-3-fold using complete RPMI 1640 medium in a 24-well plate and culturing for an additional three to five weeks.
Additionally, activated T cells were also prepared from CD19-negative cells stimulated with the human T cell activator CD3/CD28 (Dynabeads, Life Technologies, Waltham, MA, USA) according to the manufacturer’s recommendations and expanded in complete RPMI 1640 medium containing 30 U/mL recombinant IL-2 (PeproTech EC, London, UK) in a 12-well plate for three to ten days.
After successful cell expansion, both EBV-LCLs and activated T cells were harvested, and the number of viable cells was determined via trypan blue exclusion using a hemocytometer. These cells were centrifuged, resuspended at a concentration of 4-10 × 106/mL in 1 mL of cold CELLBANKER 1 plus and dispensed into two sterile 1-mL 2D tubes in 500-µL aliquots. Cryopreservation was conducted similarly to that for PBMCs.
DNA isolation
Genomic DNA was isolated from frozen buffy coat of peripheral or cord blood or from saliva in Oragene preservative solution (DNA Genotek, Otawa, Canada). An Autopure LS DNA purification system (QIAGEN, Hilden, Germany) was used for peripheral blood, and a QIAsymphony instrument (QIAGEN) was used for cord blood and saliva. Isolated DNA was suspended in 350 µL of TE buffer (10 mM Tris, 1 mM EDTA); the original concentration was measured using PicoGreen dye in a Quant-iT PicoGreen dsDNA Assay kit (Thermo Fisher Scientific). The OD260/280 ratio was measured with a microplate reader. The DNA concentration was adjusted to 50 ng/µL using an automated liquid handling system (Tecan), and the samples were stored in a Brooks SampleStore system (Brooks) at 4°C.
Quality control assays
DNA quality control (QC) assays were performed using a NanoDrop 2000 (Thermo Fisher Scientific) and an Agilent 4200 TapeStation system (Agilent Technologies, Santa Clara, CA, USA) on randomly selected DNA samples isolated between May 2013 and November 2017 (Betsou 2017). The DNA length was analyzed by pulsed field gel electrophoresis using a Bio-Rad CHEF DRII system (Bio-Rad, Hercule, CA, USA). For PBMC quality control assays, we retrospectively selected ten samples per month randomly between May 2013 and May 2017 and analyzed cell numbers using data from the CelltacF blood cell counter.
Integrated BioBank of Luxembourg-Proficiency Testing program (IBBL-PT)
The IBBL-PT is provided by IBBL, Luxembourg (https://www.ibbl.lu/ibbl-bioservices/biospecimen-proficiency-testing/). Between 2015 and 2018, we participated in three rounds of the program for cell viability, viable PBMC isolation and DNA quantification.
Results
Management of the TMM biobank
The TMM biobank is managed by the Tohoku Medical Megabank Organization (ToMMo) under the supervision of the presidents and board of directors of Tohoku University and Iwate Medical University. The establishment of the TMM biobank was financially supported by the recovery budget from the Great East Japan Earthquake; the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT); and the Japan Agency for Medical Research and Development (AMED) (Fig. 1). The TMM Project is organized by the Ethical, Legal and Social Issues (ELSI) Committee and Advisory Board as well as the Research Ethics Committee, which comprises experts both inside and outside the organization.
The biobank entrance is equipped with a biometric security system. Because Japan occasionally faces earthquakes, the TMM biobank is located in an earthquake-proof building with firewalls to protect the samples. In addition, the electronic specimen storage systems are connected to the emergency power supply system, which has the capacity to keep the freezers running for at least 72 hours.
In addition, we are keeping a part of the samples individually in different storage conditions, such as –80°C freezer and liquid nitrogen storage system in our storage facilities. As we have verified our liquid nitrogen system can keep ultra-low temperature conditions for weeks without additional liquid nitrogen and power supply, this operation helps us to maintain the quality of our samples for a long period securely against an accidental power failure.
Establishment of the TMM biobank
Recruitment of participants in the TMM cohort studies progressed as described (Kuriyama et al. 2016; Takai-Igarashi et al. 2017). The TMM CommCohort study was conducted in the Miyagi and Iwate prefectures, while the TMM BirThree cohort Study was conducted in Miyagi Prefecture. As shown in Fig. 2, we recruited CommCohort study participants from sites at which annual health examinations were conducted by municipalities and admission centers, referred to as “community support centers” (Miyagi) or “satellite centers” (Iwate). Pregnant women were recruited from obstetric clinics and hospitals for the TMM BirThree cohort study. All participants provided written informed consent, which was approved by the research ethics committees at Tohoku University Graduate School of Medicine and Iwate Medical University, to donate their deidentified specimens and data for use in a broad range of research activities performed by unspecified researchers.
Biospecimens and data collection
We collected 34 mL of blood into four distinct tubes: 1) EDTA-2Na tubes (Venoject II, VP-NA070K, Terumo, Tokyo, Japan) for plasma and buffy coat collection, 2) heparin tubes (Venoject II, VP-H050K) for PBMC collection, 3) two plain tubes with a separation gel (Venoject II, VP-P100K) for serum collection and clinical biochemical analysis, and 4) sodium fluoride tubes for blood glucose level analysis. We also collected 3-10 mL of urine from adult participants (Fig. 2). We kept records of the time of sample collection and the time of the last meal of the participants.
Notably, we collected umbilical cord blood from newborn babies into three tubes: 1) EDTA-2K tubes (BD Vacutainer® K2 EDTA tube, 365900, Becton Dickinson and Company, Franklin Lakes, NJ, USA) for plasma and buffy coat collection, 2) sodium citrate tubes with a separation gel and a density gradient liquid (BD Vacutainer® CPT™, 362761) for umbilical cord blood mononuclear cell (CBMC) collection, and 3) plain tubes with a separation gel (BD Vacutainer SSTII tube, 368972) for serum collection (Fig. 2). For older siblings (5 years or older) of the BirThree cohort participants, we collected blood (5 mL or less) and urine. For younger siblings (younger than 5 years), we collected saliva for genomic DNA analysis.
Sample numbers and transportation
During the period of initial recruitment to the TMM cohort studies, we collected biological specimens from approximately 150,000 individuals who participated in either the TMM CommCohort or the TMM BirThree cohort study. We had stored 2,799,400 tubes as of December 1, 2018 (Fig. 3A), including 1,363,200 tubes from the TMM CommCohort and 1,436,200 tubes from the TMM BirThree cohort. As of December 1, 2018, the biospecimens in the TMM biobank comprise 867,100 tubes of plasma, accounting for 25.4% of the total number of tubes; 866,400 tubes of serum (25.3%); 319,200 tubes of buffy coat (9.3%); 424,700 tubes of PBMCs and CBMCs (12.4%); 608,800 tubes of urine (17.8%); 70,300 tubes of breast milk (2.1%); and 4,600 tubes of saliva (0.1%) from the first and second surveys of the TMM cohort (Fig. 3B). The sample number excludes tubes for distribution and tubes discarded because of participant withdrawal.
We stored buffy coat, plasma, serum, urine and breast milk in the Brooks BioStore system at –80°C. In addition, we manually isolated mononuclear cells from peripheral blood or cord blood, and PBMCs and CBMCs were stored below –180°C in a liquid nitrogen storage system for long-term storage. In addition, we also established EBV-transformed B lymphocyte cell lines and activated T lymphocytes, which were also stored in the liquid nitrogen storage system (Fig. 3C).
We received biospecimens from participants in Miyagi Prefecture in the late afternoon to early evening and generally processed and stored these samples on the day of collection. However, biospecimens from participants in Iwate Prefecture were received and processed on the next day (Fig. 3D). Blood in blood collection tubes, urine and breast milk in centrifuge tubes, and saliva in Oragene (Oragene OG-500) tubes were shipped in temperature-controlled boxes. We have stored saliva samples at 4°C in the Oragene, thereby keeping DNAs stably even above 0°C. The overall procedures of the TMM biobank are controlled by the LIMS, and numerous automated systems have been exploited for liquid specimen handling, DNA extraction, and autostorage (+4°C and –80°C).
Collection of the samples
The increase in the number of stored biospecimen tubes reflected a seasonal change in the recruitment of the CommCohort participants, peaking between May and September. This pattern arose because we recruited the CommCohort participants partially in collaboration with the annual health examinations by municipalities, which are generally conducted in the spring and summer. In contrast, the number of BirThree cohort participants increases constantly throughout the seasons, as these participants were recruited in Obstetrics and Gynecology clinics and hospitals at the beginning of pregnancy (Fig. 4A). We instituted a second-phase survey of TMM cohort participants, which started on June 1, 2017. Thus, the number of collected tubes has been increasing continuously (Fig. 4B). In the second-phase survey, we have been collecting samples mainly in the community support centers and satellite centers. Therefore, the increase in the number of collected samples is relatively stable (Fig. 4A). Notably, we received samples from more than 700 individuals on July 1, 2015, which is the highest number of biospecimens we have ever processed.

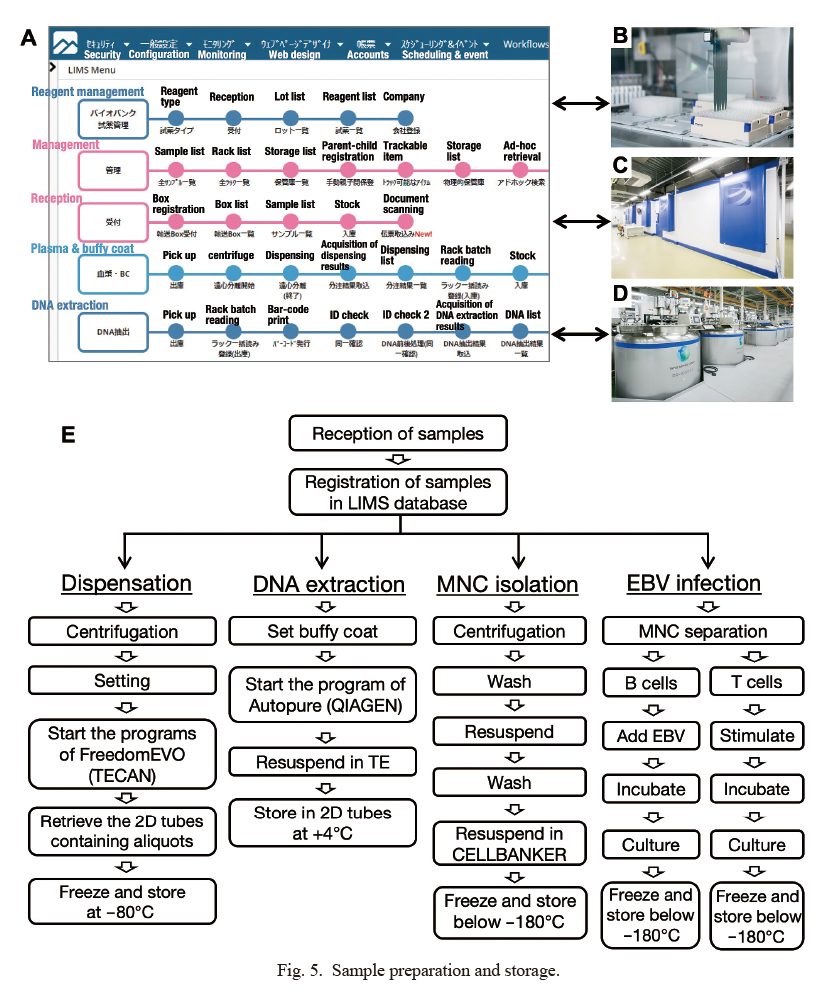
We received deidentified samples of blood and other biological fluids, which were then processed and stored in our biobank systems. All procedures in our biobank are managed by a single customized LIMS (Fig. 5A), which connects the barcode readers, automatic liquid aliquoting systems (Fig. 5B), DNA extraction systems, automatic storage systems (Fig. 5C) and liquid nitrogen storage systems (Fig. 5D). As summarized in Fig. 5E, these multiprocessing systems are controlled elaborately and closely by the LIMS in a closed, firewall-protected network (Takai-Igarashi et al. 2017).
The links between the deidentified participant ID and storage tube ID, timestamp, specimen location, liquid volume, and cell counts are also controlled by the LIMS. In addition, we input quality-related information, such as the cell culture passage number and lack of sufficient quality caused by nonconforming events into the LIMS. We restrict the access to the LIMS based on the assigned roles of staff members, such as sample registration, EBV-LCL establishment, and DNA purification (Fig. 5E). Using a combination of the LIMS and automation systems, we can process biospecimens obtained from as many as 700 individuals per day.
DNA isolation and quality
We have isolated genomic DNA from the buffy coat of peripheral blood and cord blood from 136,500 participants (as of December 1, 2018). From the buffy coat layer, corresponding to 4.9 mL of peripheral blood, we obtained an average of 50.1 ± 23.6 µg (mean ± SD) of double-stranded DNA per person between May 2013 and Dec 2017, as measured with the PicoGreen system. To assess the DNA quality, we analyzed 118 randomly chosen specimens obtained from May 2013 to Dec 2017 on a TapeStation. The average DNA size of these samples was greater than 50 kb, and the DNA integrity number (DIN) was 9.2 ± 0.4 (Fig. 6A, B). We also conducted pulsed field gel electrophoresis. The length of our DNA specimens was greater than 50 kb, which assured that our DNA samples are appropriate for use in long-read genomic DNA sequencing (Fig. 6C). In addition, we measured the OD260/280 ratio of the DNA samples via a microplate reader equipped with the Tecan system; the average ratio was 1.77 ± 0.09 (n = 23,004). These results thus indicate that the TMM biobank has extracted a reasonable amount of DNA for data sharing and that the quality of the DNA samples is of a sufficiently high standard to satisfy criteria for use in even long-read genomic DNA sequencing.

TMM is conducting multilayered omics analyses. The TMM biobank exploits plasma samples instead of serum for metabolomic and proteomic analyses (Koshiba et al. 2018). The preanalytical condition of samples greatly affects the metabolomic results. We retain the records of the working temperature and elapsed time in the LIMS for all procedures from specimen receipt to storage. These records allow users to select suitable specimens for analysis. Notably, the association of single nucleotide polymorphisms with plasma metabolite concentrations has been analyzed, and the results are available (Tadaka et al. 2018). Currently, the summarized data for the metabolomic (n = 10,000) and proteomic (n = 500) analyses are on our website, called jMorp (https://jmorp.megabank.tohoku.ac.jp/2017/compounds). These results thus show that the plasma and serum samples from the TMM biobank are well organized and suitable for use in advanced metabolomic and proteomic analyses.
PBMC and CBMC isolation and EBV-LCL/activated T cell establishment
We isolated PBMCs and CBMCs manually from most participants. In our processing procedure, the average yield of PBMCs was 7.4 ± 3.2 × 106 cells (n = 142,368) from 5 mL of peripheral blood, as analyzed via the blood cell counter.
The TMM biobank establishes EBV-LCLs and activated T cells. As of December 1, 2018, we had established EBV-LCLs from 2,987 participants’ PBMCs and stored activated T cells from 3,467 participants’ PBMCs. We selected the participants for the establishment of LCLs based on the availability of whole-genome sequence data (Yamaguchi-Kabata et al. 2015, Yasuda et al. 2019). We achieved an efficiency of 85.1% for the establishment of EB-LCLs and 99.8% for the establishment of activated T cells. We minimized human error during the procedure by the use of the customized LIMS.
Standardization of biobank quality control
Since 2015, we have been participating annually in the IBBL-PT (Gaignaux et al. 2016), which is endorsed by the International Society for Biological and Environmental Repositories (ISBER). We participated in the tests for PBMC isolation, cell viability and DNA quantification. The IBBL-PT results indicate that the quality of our specimens is comparable to those from other IBBL-PT participants worldwide.
Biobanks require reliable quality control for sample collection, processing, and storage. To systematically and effectively manage quality improvement activities, the Group of Biobank at TMM acquired International Organization for Standardization (ISO) 9001:2008 certification for quality management in June 2015. Furthermore, in March 2016, the Group of Biobank, Group of Integrated Database Systems, and Group of Materials and Information Management received ISO 27001:2013 certification (ISO/IECJTC1/SC27 2013) for information security. These three groups are currently certified under both ISO 9001:2015 (ISO/TC176/SC2 2015) and ISO 27001:2013.
In addition, to eliminate the possibility of sample mix-up and ID mislabeling, we confirm the collation of sex and ABO blood type data (Lang et al. 2016), including cohort questionnaire data, ABO blood cell typing in EDTA blood collection tubes, and genomic data. To confirm and correct mismatches, we established a system that enables re-extraction of DNA from backup buffy coat specimens and performs quantitative real-time PCR and/or MassARRAY analysis (Gabriel et al. 2009) of our DNA specimens.
Sample and data sharing
An important activity of the TMM biobank is sample and data sharing. To this end, we established the dbTMM (Yasuda et al. 2019). The catalog of available resources and the application processes are provided on the dbTMM website (http://www.dist.megabank.tohoku.ac.jp/index.html, Japanese only). We started the sample and data sharing program in August 2015. Each applicant is reviewed and approved by the Sample and Data Access Committee. The members of this committee include genomic research experts, lawyers, and community resident representatives outside TMM. As of the end of December 2018, 23 data sharing projects and over 100 collaboration projects had been approved. Details of our sample and data sharing procedures and activities will be reported separately.
Discussion
The TMM biobank has collected various biospecimens and associated data from approximately 150,000 participants. In the first survey, from 2013 to 2017, we managed more than 2.8 million tubes containing biospecimens. To handle this large number of tubes safely, properly and efficiently, we constructed and incorporated several automation systems into our biobank. Furthermore, to reduce human errors and trace biobank procedures, we customized a LIMS and integrated information for the automated sample dispensing, isolation and storage systems as well as for the sample identity confirmation system. Since all tubes, tube racks, and positions in the storage units are registered and linked in the LIMS, we can select and collect tubes in response to various biospecimen sharing requests. We believe that these biobank systems generate an important infrastructure for the realization of next-generation medicine.
The TMM biobank has been certified by the ISO 9001 quality management system and the ISO 27001 information security management system. These two certifications show our commitment to high-quality and robust information security. As a result, we have improved the reliability of the organization and strengthened its competitiveness. Moreover, notably our DNA samples have a length of greater than 50 kbp. The length and purity of our DNA specimens are sufficient for various types of usage, including long-read genomic DNA sequencing, by using PacBio systems (Huddleston et al. 2017) or even the Nanopore MinION sequencing system (Feng et al. 2015).
The TMM biobank stores PBMCs and EBV-LCLs/activated T cells. In our routine practice, an average of 7.6 × 106 cells are recovered from 5 mL of peripheral blood, which is comparable to reported results (Fuss et al. 2009). PBMCs are one of the most important cells in the immune system because they are responsible for pathogen defense and for the maintenance of homeostasis in the body (Kersey and Gajl-Peczalska 1975). Therefore, PBMCs are often used in various immunological and toxicological studies (Santovito et al. 2014; Michalowicz et al. 2013). Notably, PBMCs have also been used recently for personal omics profiling studies aiming to reveal dynamic molecular and medical phenotypes (Chen et al. 2012). The TMM biobank considers EBV-LCLs to be the most reliable yet low-cost cells. EBV-LCLs are a convenient representation of cells from unrelated individuals for in vitro cell culture research projects.
After cell line establishment, continuous growth of EBV-LCLs in vitro can be maintained via relatively simple procedures. Moreover, the genomes of these cells remain stable during subsequent cell divisions, in contrast to those of human cancer cell lines, which undergo frequent genetic rearrangements (Londin et al. 2011; Nickles et al. 2012). Because large quantities of DNA are needed for long-read genome sequencing, we consider biobank-originated EBV-LCLs to be an important resource for the realization of genomic medicine. In fact, EBV-LCLs are sometimes referred to as personalized cell lines (Tilgner et al. 2014). Whereas some epigenetic signatures may be lost during the immortalization process of EBV-LCLs, others may remain intact even after the establishment of EBV-LCLs and may aid the discovery of disease-related epigenomic modifications (Nguyen et al. 2010). Therefore, EBV-LCLs may be applicable for multiomic analyses. Personalized EBV-LCLs may also be a valuable research tool for analyzing the efficacy and/or adverse reactions of drug treatment (Niu and Wang 2015). We surmise that PBMCs, EBV-LCLs and activated T cells are also important resources for the generation of human induced pluripotent stem cells (iPS cells), which represent another type of personalized cell line used as a research tool in personalized medicine.
Several disease cohort biobanks have been established in Japan (Nagai et al. 2017). In this regard, the TMM biobank is the biobank that aims to investigate the relationships among the earthquake exposure level, psychological impact and long-term health effects via earthquake-related questionnaires. However, the most important feature of the TMM biobank is that it is the largest population-based biobank in Japan, covering a wide range of generations. We earnestly anticipate that specimens from the TMM biobank will contribute as powerful tools for identifying disease-related genes and seeking disease-related biomarkers in the Japanese population. Similarly, while many genome reference panels have been constructed worldwide, such as the 1000 Genomes Project (1000 Genomes Project Consortium et al. 2015) and ClinVar (Landrum et al. 2016), a suitable reference background is required to increase the accuracy of case-control studies, because the ethnicity of the Japanese population is clearly different from those underlying the available reference panels (Nagasaki et al. 2015; Yasuda et al. 2018). We finished the TMM cohort initial survey and have started the second survey in June 2017 to explore health and growth status of the participants in TMM ComCohort and TMM BirThree cohorts, which will help to acquire longitudinal human health data. We believe that our cohort research and biobank systems will be an important infrastructure to actualize next-generation personalized medicine and healthcare.
Acknowledgments
The TMM biobank was supported as part of the reconstruction from the Great East Japan Earthquake on March 11, 2011 by the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT) and the Japan Agency for Medical Research and Development (AMED) (grant numbers JP17km0105001 and JP17km0105002). The authors appreciate all the participants in the TMM cohorts and the staff of the TMM projects. The ToMMo supercomputer system (http://sc.megabank.tohoku.ac.jp/en) is supported by the Facilitation of the R&D Platform for AMED Genome Medicine Support conducted by AMED (grant number JP17km0405001). We would especially like to thank the members of the Department of Biobank Lifescience, for their technical assistance with the establishment of and sample processing at the TMM biobank.
Conflict of Interest
The authors declare no conflict of interest.
References
-
1000 Genomes Project Consortium,
Auton, A.,
Brooks,
L.D.,
Durbin,
R.M.,
Garrison,
E.P.,
Kang,
H.M.,
Korbel,
J.O.,
Marchini,
J.L.,
McCarthy,
S.,
McVean,
G.A. &
Abecasis,
G.R.
(2015) A global reference for human genetic variation. Nature, 526, 68-74.
-
Betsou,
F.
(2017) Quality assurance and quality control in biobanking. In Biobanking of human biospecimens: principles and practice, edited by Hainaut, P., Vaught, J., Zatloukal, K. & Pasterk, M. Springer International Publishing, Cham, pp. 23-49.
-
Chen,
R.,
Mias,
G.I.,
Li-Pook-Than,
J.,
Jiang,
L.,
Lam,
H.Y.,
Chen,
R.,
Miriami,
E.,
Karczewski,
K.J.,
Hariharan,
M.,
Dewey,
F.E.,
Cheng,
Y.,
Clark,
M.J.,
Im,
H.,
Habegger,
L.,
Balasubramanian,
S.,
et al. (2012) Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell, 148, 1293-1307.
-
Feng,
Y.,
Zhang,
Y.,
Ying,
C.,
Wang,
D. &
Du,
C.
(2015) Nanopore-based fourth-generation DNA sequencing technology. Genomics Proteomics Bioinformatics, 13, 4-16.
-
Fuss,
I.J.,
Kanof,
M.E.,
Smith,
P.D. &
Zola,
H.
(2009) Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr. Protoc. Immunol., 85, 7.1.1-7.1.8.
-
Gabriel,
S.,
Ziaugra,
L. &
Tabbaa,
D.
(2009) SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr. Protoc. Hum. Genet., 60, 2.12.1-2.12.18.
-
Gaignaux,
A.,
Ashton,
G.,
Coppola,
D.,
De Souza,
Y.,
De Wilde,
A.,
Eliason,
J.,
Grizzle,
W.,
Guadagni,
F.,
Gunter,
E.,
Koppandi,
I.,
Shea,
K.,
Shi,
T.,
Stein,
J.A.,
Sobel,
M.E.,
Tybring,
G.,
et al. (2016) A biospecimen proficiency testing program for biobank accreditation: four years of experience. Biopreserv. Biobank., 14, 429-439.
-
Huddleston,
J.,
Chaisson,
M.J.P.,
Steinberg,
K.M.,
Warren,
W.,
Hoekzema,
K.,
Gordon,
D.,
Graves-Lindsay,
T.A.,
Munson,
K.M.,
Kronenberg,
Z.N.,
Vives,
L.,
Peluso,
P.,
Boitano,
M.,
Chin,
C.S.,
Korlach,
J.,
Wilson,
R.K.,
et al. (2017) Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res., 27, 677-685.
-
ISO/IECJTC1/SC27
(2013) ISO/IEC 27001:2013 Information technology: Security techniques: Information security management systems: Requirements. https://www.iso.org/standard/54534.html [Accessed: March 8, 2019]
-
ISO/TC176/SC2
(2015) ISO 9001:2015 Quality management systems: Requirements. https://www.iso.org/standard/62085.html [Accessed: March 8, 2019]
-
Kersey,
J.H. &
Gajl-Peczalska,
J.
(1975) T and B lymphocytes in humans. A review. Am. J. Pathol., 81, 446-458.
-
Koshiba,
S.,
Motoike,
I.,
Kojima,
K.,
Hasegawa,
T.,
Shirota,
M.,
Saito,
T.,
Saigusa,
D.,
Danjoh,
I.,
Katsuoka,
F.,
Ogishima,
S.,
Kawai,
Y.,
Yamaguchi-Kabata,
Y.,
Sakurai,
M.,
Hirano,
S.,
Nakata,
J.,
et al. (2016) The structural origin of metabolic quantitative diversity. Sci. Rep., 6, 31463.
-
Koshiba,
S.,
Motoike,
I.,
Saigusa,
D.,
Inoue,
J.,
Shirota,
M.,
Katoh,
Y.,
Katsuoka,
F.,
Danjoh,
I.,
Hozawa,
A.,
Kuriyama,
S.,
Minegishi,
N.,
Nagasaki,
M.,
Takai-Igarashi,
T.,
Ogishima,
S.,
Fuse,
N.,
et al. (2018) Omics research project on prospective cohort studies from the Tohoku Medical Megabank Project. Genes Cells, 23, 406-417.
-
Kuriyama,
S.,
Yaegashi,
N.,
Nagami,
F.,
Arai,
T.,
Kawaguchi,
Y.,
Osumi,
N.,
Sakaida,
M.,
Suzuki,
Y.,
Nakayama,
K.,
Hashizume,
H.,
Tamiya,
G.,
Kawame,
H.,
Suzuki,
K.,
Hozawa,
A.,
Nakaya,
N.,
et al. (2016) The Tohoku Medical Megabank Project: design and mission. J. Epidemiol., 26, 493-511.
-
Landrum,
M.J.,
Lee,
J.M.,
Benson,
M.,
Brown,
G.,
Chao,
C.,
Chitipiralla,
S.,
Gu,
B.,
Hart,
J.,
Hoffman,
D.,
Hoover,
J.,
Jang,
W.,
Katz,
K.,
Ovetsky,
M.,
Riley,
G.,
Sethi,
A.,
et al. (2016) ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res., 44, D862-868.
-
Lang,
K.,
Wagner,
I.,
Schone,
B.,
Schofl,
G.,
Birkner,
K.,
Hofmann,
J.A.,
Sauter,
J.,
Pingel,
J.,
Bohme,
I.,
Schmidt,
A.H. &
Lange,
V.
(2016) ABO allele-level frequency estimation based on population-scale genotyping by next generation sequencing. BMC Genomics, 17, 374.
-
Londin,
E.R.,
Keller,
M.A.,
D’Andrea,
M.R.,
Delgrosso,
K.,
Ertel,
A.,
Surrey,
S. &
Fortina,
P.
(2011) Whole-exome sequencing of DNA from peripheral blood mononuclear cells (PBMC) and EBV-transformed lymphocytes from the same donor. BMC Genomics, 12, 464.
-
Mathay,
C.,
Ammerlaan,
W. &
Betsou,
F.
(2012) Automatic buffy coat extraction: methodology, feasibility, optimization and validation study. Biopreserv. Biobank., 10, 543-547.
-
Michalowicz,
J.,
Mokra,
K.,
Rosiak,
K.,
Sicinska,
P. &
Bukowska,
B.
(2013) Chlorobenzenes, lindane and dieldrin induce apoptotic alterations in human peripheral blood lymphocytes (in vitro study). Environ. Toxicol. Pharmacol., 36, 979-988.
-
Nagai,
A.,
Hirata,
M.,
Kamatani,
Y.,
Muto,
K.,
Matsuda,
K.,
Kiyohara,
Y.,
Ninomiya,
T.,
Tamakoshi,
A.,
Yamagata,
Z.,
Mushiroda,
T.,
Murakami,
Y.,
Yuji,
K.,
Furukawa,
Y.,
Zembutsu,
H.,
Tanaka,
T.,
et al. (2017) Overview of the biobank Japan project: study design and profile. J. Epidemiol., 27, S2-S8.
-
Nagasaki,
M.,
Yasuda,
J.,
Katsuoka,
F.,
Nariai,
N.,
Kojima,
K.,
Kawai,
Y.,
Yamaguchi-Kabata,
Y.,
Yokozawa,
J.,
Danjoh,
I.,
Saito,
S.,
Sato,
Y.,
Mimori,
T.,
Tsuda,
K.,
Saito,
R.,
Pan,
X.,
et al. (2015) Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat. Commun., 6, 8018.
-
National Police Agency
(2019) Police countermeasures and damage situation associated with 2011 Tohoku district - off the Pacific Ocean Earthquake. https://www.npa.go.jp/news/other/earthquake2011/pdf/higaijokyo_e.pdf [Accessed: March 8, 2019]
-
Neitzel,
H.
(1986) A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum. Genet., 73, 320-326.
-
Nguyen,
A.,
Rauch,
T.A.,
Pfeifer,
G.P. &
Hu,
V.W.
(2010) Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB J., 24, 3036-3051.
-
Nickles,
D.,
Madireddy,
L.,
Yang,
S.,
Khankhanian,
P.,
Lincoln,
S.,
Hauser,
S.L.,
Oksenberg,
J.R. &
Baranzini,
S.E.
(2012) In depth comparison of an individual’s DNA and its lymphoblastoid cell line using whole genome sequencing. BMC Genomics, 13, 477.
-
Niu,
N. &
Wang,
L.
(2015) In vitro human cell line models to predict clinical response to anticancer drugs. Pharmacogenomics, 16, 273-285.
-
Saigusa,
D.,
Okamura,
Y.,
Motoike,
I.N.,
Katoh,
Y.,
Kurosawa,
Y.,
Saijyo,
R.,
Koshiba,
S.,
Yasuda,
J.,
Motohashi,
H.,
Sugawara,
J.,
Tanabe,
O.,
Kinoshita,
K. &
Yamamoto,
M.
(2016) Establishment of protocols for global metabolomics by LC-MS for biomarker discovery. PLoS One, 11, e0160555.
-
Santovito,
A.,
Cervella,
P. &
Delpero,
M.
(2014) Chromosomal damage in peripheral blood lymphocytes from nurses occupationally exposed to chemicals. Hum. Exp. Toxicol., 33, 897-903.
-
Tadaka,
S.,
Saigusa,
D.,
Motoike,
I.N.,
Inoue,
J.,
Aoki,
Y.,
Shirota,
M.,
Koshiba,
S.,
Yamamoto,
M. &
Kinoshita,
K.
(2018) jMorp: Japanese multi omics reference panel. Nucleic Acids Res., 46, D551-D557.
-
Takai-Igarashi,
T.,
Kinoshita,
K.,
Nagasaki,
M.,
Ogishima,
S.,
Nakamura,
N.,
Nagase,
S.,
Nagaie,
S.,
Saito,
T.,
Nagami,
F.,
Minegishi,
N.,
Suzuki,
Y.,
Suzuki,
K.,
Hashizume,
H.,
Kuriyama,
S.,
Hozawa,
A.,
et al. (2017) Security controls in an integrated biobank to protect privacy in data sharing: rationale and study design. BMC Med. Inform. Decis. Mak., 17, 100.
-
Tilgner,
H.,
Grubert,
F.,
Sharon,
D. &
Snyder,
M.P.
(2014) Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc. Natl. Acad. Sci. USA, 111, 9869-9874.
-
Yamaguchi-Kabata,
Y.,
Nariai,
N.,
Kawai,
Y.,
Sato,
Y.,
Kojima,
K.,
Tateno,
M.,
Katsuoka,
F.,
Yasuda,
J.,
Yamamoto,
M. &
Nagasaki,
M.
(2015) iJGVD: an integrative Japanese genome variation database based on whole-genome sequencing. Hum. Genome Var., 2, 15050.
-
Yasuda,
J.,
Katsuoka,
F.,
Danjoh,
I.,
Kawai,
Y.,
Kojima,
K.,
Nagasaki,
M.,
Saito,
S.,
Yamaguchi-Kabata,
Y.,
Tadaka,
S.,
Motoike,
I.N.,
Kumada,
K.,
Sakurai-Yageta,
M.,
Tanabe,
O.,
Fuse,
N.,
Tamiya,
G.,
et al. (2018) Regional genetic differences among Japanese populations and performance of genotype imputation using whole-genome reference panel of the Tohoku Medical Megabank Project. BMC Genomics, 19, 551.
-
Yasuda,
J.,
Kinoshita,
K.,
Katsuoka,
F.,
Danjoh,
I.,
Sakurai-Yageta,
M.,
Motoike,
I.N.,
Kuroki,
Y.,
Saito,
S.,
Kojima,
K.,
Shirota,
M.,
Saigusa,
D.,
Otsuki,
A.,
Kawashima,
J.,
Yamaguchi-Kabata,
Y.,
Tadaka,
S.,
et al. (2019) Genome analyses for the Tohoku Medical Megabank Project toward establishment of personalized healthcare. J. Biochem., 165, 139-158.