2021 Volume 254 Issue 3 Pages 233-243
2021 Volume 254 Issue 3 Pages 233-243
Prognosis of patients with hepatocellular carcinoma remains poor because of progression of hepatocellular carcinoma and high recurrence rates. Cyclin D2 (CCND2) plays a vital role in regulating the cell cycle; indeed, aberrant methylation of CCND2 is involved in the development of hepatocellular carcinoma. Therefore, we aimed to investigate levels of CCND2 methylation in patients with hepatitis B virus (HBV)-associated hepatocellular carcinoma and to evaluate its prognostic significance after hepatectomy. In total, 257 subjects were enrolled (166 hepatocellular carcinoma patients undergoing surgical resection, 61 chronic hepatitis B (CHB) patients, and 30 healthy controls). CCND2 methylation in peripheral blood mononuclear cells was measured quantitatively using MethyLight. We found that CCND2 methylation levels in patients with HBV-associated hepatocellular carcinoma were significantly higher than in CHB patients (P < 0.001) or healthy controls (P < 0.001). Within the hepatocellular carcinoma group, CCND2 methylation levels were higher in patients with portal vein invasion, early tumor recurrence, TNM III/IV stage, and tumor size ≥ 5 cm (P < 0.05). Furthermore, higher levels of CCND2 methylation were associated with worse overall survival and disease-free survival (P = 0.005 and P < 0.001, respectively). Multivariate analysis identified CCND2 methylation as an independent prognostic factor for early tumor recurrence (P = 0.021), overall survival (P = 0.022), and disease-free survival (P < 0.001) in hepatocellular carcinoma patients after resection. In conclusion, hypermethylation of CCND2 may have clinical utility for predicting a high risk of poor prognosis and early tumor recurrence in patients with HBV-associated hepatocellular carcinoma after hepatectomy.
Hepatocellular carcinoma accounts for the majority of primary liver cancers. Indeed, it is the second leading cause of cancer-related mortality in men worldwide and the incidence is increasing (Torre et al. 2015; Yang et al. 2019). In China, the main cause of hepatocellular carcinoma is chronic infection with hepatitis B virus (HBV) (Yuen et al. 2009; Torre et al. 2015). Although new surgical techniques have improved the survival of patients with hepatocellular carcinoma, some still have undesirable outcomes; indeed, the 5-year median survival rate is approximately 50% (Takayama 2011). This poor prognosis is due mainly to intrahepatic metastases or post-surgical recurrence, particularly intrahepatic recurrence. Therefore, it is essential to identify a robust biomarker capable of predicting the prognosis of hepatocellular carcinoma and guiding subsequent treatment.
DNA methylation is a major epigenetic alteration that occurs generally in the CpG islands of gene promoter regions; such methylations can trigger aberrant gene expression. DNA methyltransferases add a methyl group to the 5-position of cytosine (Baylin et al. 1998; Herman and Baylin 2003). Similar to other tumors, carcinogenesis and progression of hepatocellular carcinoma are driven by complex genetic and epigenetic alterations. Several studies show that cancer-linked DNA methylation is closely related to hepatocellular carcinoma initiation, progression, and invasion (Liu et al. 2017; Tian et al. 2017).
Cyclin D2 (CCND2) is a member of the D-type cyclin family. In addition to promoting cell proliferation, it has growth-inhibitory effects. Aberrant expression of CCND2 arrests cell cycle progression, suggesting that CCND2 inhibits cell proliferation (Tam et al. 1994; Meyyappan et al. 1996; Meyyappan et al. 1998). This may be circumvented by downregulation of CCND2 expression via promoter hypermethylation, a common factor in hepatocellular carcinoma (Wang et al. 2012).
Here, we analyzed the methylation status of the CCND2 promoter in patients with hepatocellular carcinoma using MethyLight, a high-throughput assay that measures DNA methylation. More importantly, we explored the utility of CCND2 methylation for predicting prognosis of patients undergoing hepatectomy as a treatment for HBV-associated hepatocellular carcinoma.
The study enrolled 257 subjects attending the Qilu Hospital of Shandong University between February 2013 and April 2016. The cohort comprised 166 consecutive hepatocellular carcinoma patients undergoing surgical resection, 61 patients with chronic hepatitis B (CHB), and 30 healthy controls. All hepatocellular carcinoma patients were HBsAg-positive according to the 2010 update of the American Association for the Study of Liver Diseases Practice Guidelines for Management of Hepatocellular Carcinoma (Bruix et al. 2011). All patients were followed up from the date of surgery to March 2019. Early tumor recurrence was defined as recurrence within 1 year after curative hepatectomy (Yuan et al. 2007; Tsutsui et al. 2010). The study protocol was approved by the Medical Ethical Committee of Qilu Hospital of Shandong University, and all patients provided written informed consent.
Isolation of peripheral blood mononuclear cells (PBMCs)Briefly, citrate-anticoagulated peripheral blood (5 mL) was collected from all subjects. PBMCs were collected after density gradient centrifugation of blood samples on Ficoll-Paque (GE Healthcare, Uppsala, Sweden). Cells were stored at −80°C until required.
DNA extraction and TaqMan probe-based quantitative methylation-specific PCR (MethyLight)DNA was extracted from PBMCs using the QIAamp DNA Blood Mini Kit (Qiagen, Mainz, Germany), followed by bisulfite treatment using an EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA). Finally, 20 μL of modified DNA was acquired and stored at −20°C for later use.
Methylation of the CCND2 promoter was detected using MethyLight. The primer pairs and probe used to target CCND2 and the control gene (β-actin) were described previously (Feng et al. 2008); the sequences are shown in Table 1. MethyLight was carried out in a volume of 10 μL, comprising 2 μL nuclease-free water, 5 μL MethyLight Master Mix (HotStarTaq Plus DNA Polymerase, EpiTect Probe PCR Buffer and dNTP mix), 0.2 μL of TaqMan probe, 0.4 μL of each forward and reverse primers, and 2 μL bisulfite-converted DNA. The MethyLight assay was performed in an Agilent Technologies Stratagene Mx3005P apparatus (Stratagene, La Jolla, CA, USA) using a standard PCR protocol: 95°C for 5 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. DNA treated with M.SssI (Qiagen, Hilden, Germany) served as a fully methylated reference. MethyLight data were expressed as the percentage methylated reference (PMR) value, which was calculated as follows:
PMR = 100% × 2 exp [Delta Ct (target gene in sample − control gene in sample) − Delta Ct (100% methylated target gene in reference sample − control gene in reference sample)] (Wu et al. 2012).

List of primers and probes used in the study.
CCND2, cyclin D2; RT-qPCR, quantitative real-time polymerase chain reaction.
Total RNA was extracted from PBMCs using the phenol chloroform isopropanol method. Extracted RNA was resuspended in 20 μL of nuclease-free water and reverse transcribed into complementary DNA (cDNA) using the PrimerScript™ RT Reagent Kit (Takara, Shiga, Japan). CCND2 mRNA was detected by RT-qPCR using SYBR Green PCR mix (Takara). Reactions were carried out in an Agilent Technologies Stratagene Mx3005P apparatus (Stratagene, La Jolla, CA, USA). The endogenous control was β-actin. The primer sequences have been described (Fan et al. 2016; Tsutsui et al. 2010) (Table 1). The PCR protocol was as follows: 95°C for 30 s, followed by 40 cycles at 95°C for 5 s, 59°C for 30 s, and a final step of 72°C for 30 s. All PCR products were quantified using the comparative (2-∆Ct) method.
Statistical analysisStatistical analyses were performed using SPSS version 17.0 software (SPSS Inc., Chicago, IL, USA). The Mann-Whitney U-test was used to compare quantitative variables. The Chi-square test was used to compare categorical variables. Correlations between variables were analyzed using Spearman’s correlation. Survival time was determined using the Kaplan-Meier method and the results were compared using the log-rank test. Multivariate analyses were performed using Cox proportional hazards models to identify prognostic factors that predict overall survival (OS), disease-free survival (DFS), and early tumor recurrence. A P value < 0.05 was considered significant.
We used MethyLight to examine methylation levels of CCND2 promoter in hepatocellular carcinoma, CHB and healthy control groups. The MethyLight data were expressed as the PMR. Methylation map showing distribution of the PMR values of CCND2 gene in different groups of participants was presented in Fig. 1.
As shown in Fig. 2, the methylation levels of CCND2 in patients with HBV-associated hepatocellular carcinoma (median, 27.26%; interquartile range, 14.41-52.76%) were significantly higher than that in CHB patients (median, 5.33%; interquartile range, 2.38-10.33%, P < 0.001) and healthy controls (median, 4.59%; interquartile range 2.83-6.60%, P< 0.001). There was no significant difference in methylation levels between patients with CHB and healthy controls (P > 0.05). Furthermore, Spearman’s rank correlation analysis revealed a negative correlation between CCND2 methylation and CCND2 mRNA expression in patients with hepatocellular carcinoma (r = −0.46, P < 0.001; Fig. 3).
To evaluate the clinicopathological significance of CCND2 methylation, we explored the association between CCND2 methylation levels and the clinicopathological characteristics of patients with hepatocellular carcinoma (Fig. 4). As shown in Fig. 4A-D, CCND2 methylation levels in hepatocellular carcinoma patients with portal vein invasion (median, 39.78%; interquartile range, 22.93-70.96%) were significantly higher than in patients without vascular invasion (median, 26.42%; interquartile range, 12.54-43.83%; P < 0.01), higher in patients with early tumor recurrence (median, 54.71%; interquartile range, 24.15-72.70%) than in those without early tumor recurrence (median, 22.69%; interquartile range, 12.24-39.50%, P < 0.001), higher in patients with TNM III/IV stage (median, 39.78%; interquartile range, 22.93-70.96%) than in those with TNM I/II stage (median, 23.00%; interquartile range, 11.99-40.34%, P < 0.001), and higher in patients with tumor size ≥ 5 cm (median, 30.57%; interquartile range, 19.35-55.29%) than in those with tumor size < 5 cm (median, 21.40%; interquartile range, 12.12-43.83%, P = 0.013).
Thus, CCND2 methylation correlated with portal vein invasion, early tumor recurrence, TNM stage, and tumor size in patients with hepatocellular carcinoma, indicating that CCND2 methylation might be involved in tumor progression. However, there was no significant correlation between CCND2 methylation levels and other clinicopathological parameters (Fig. 4E-L).

Methylation map of the percentage methylated reference (PMR) values for cyclin D2 (CCND2) gene in different groups of participants.
HCC, hepatocellular carcinoma; CHB, chronic hepatitis B; HCs, healthy controls.

Methylation levels of cyclin D2 (CCND2) in patients with hepatitis B virus (HBV)-associated hepatocellular carcinoma (HCC), patients with chronic hepatitis B (CHB), and healthy controls (HCs).
The methylation levels of CCND2 in patients with HBV-associated HCC (median, 27.26%; interquartile range, 14.41-52.76%) were significantly higher than that in patients with CHB (median, 5.33%; interquartile range, 2.38-10.33%, P < 0.001) or in HCs (median 4.59%, interquartile range, 2.83-6.60%, P < 0.001). ***P < 0.001.

Correlation between cyclin D2 (CCND2) methylation levels and CCND2 mRNA expression in the hepatocellular carcinoma (HCC) group.
CCND2 methylation levels correlated negatively with CCND2 expression in patients with HCC (r = −0.46, P < 0.001).

Association between cyclin D2 (CCND2) promoter methylation levels and clinicopathological parameters of patients with hepatocellular carcinoma (HCC).
(A) The percentage methylated reference (PMR) value of CCND2 was significantly higher in HCC patients with portal vein invasion (median, 39.78%) than in those without (median, 26.42%; P < 0.01).
(B) The PMR value of CCND2 was significantly higher in patients with early tumor recurrence (median, 54.71%) than in those without (median, 22.69%; P < 0.001).
(C) The PMR value of CCND2 was significantly higher in patients with TNM III/IV stage (median, 39.78%) than in those with TNM I/II stage (median, 23.00%; P < 0.001).
(D) The PMR value of CCND2 was significantly higher in patients with tumor size ≥ 5 cm (median, 30.57%) than in those with tumor size < 5 cm (median, 21.40%; P = 0.013).
(E-L) There was no significant correlation between the PMR value of CCND2 and gender, age, HBsAg, hepatitis B virus (HBV) DNA, histological grading, liver cirrhosis, tumor number, or alpha-fetoprotein (AFP) level.
*** P < 0.001, ** P < 0.01, and * P < 0.05.
Next, we constructed Kaplan-Meier curves to examine relationships between CCND2 methylation and outcomes of hepatocellular carcinoma patients. We selected a median PMR value of 27.26% as the cutoff point for distinguishing high and low CCND2 methylation levels. Thus, among the 166 hepatocellular carcinoma patients examined, 83 with a PMR > 27.26% were defined as the higher methylation level group; patients with a PMR ≤ 27.26% were defined as the lower methylation level group. As shown in Fig. 5, OS in the higher methylation level group was significantly shorter than that in the lower methylation group (P = 0.005). In addition, patients with higher CCND2 methylation levels had poorer DFS (P < 0.001) than those with lower CCND2 methylation levels.
Next, we used univariate and multivariate analyses to identify factors prognostic for OS and DFS. Univariate analyses identified tumor number, tumor size, portal vein invasion, TNM stage, and CCND2 methylation level as being associated with OS (Table 2). All five variables were entered into multivariate analyses using Cox proportional hazards models. The results identified TNM stage (hazard ratio (HR), 3.520; 95% confidence interval (CI), 1.102-11.240; P = 0.034) and high CCND2 methylation levels (HR, 1.740; 95% CI, 1.082-2.799; P = 0.022) as independent risk factors for OS (Table 3).
Moreover, univariate analyses identified tumor number, tumor size, portal vein invasion, TNM stage, and CCND2 methylation level as being significantly associated with DFS (Table 4). Multivariate analyses identified only CCND2 methylation level (HR, 2.184; 95% CI, 1.472-3.240; P < 0.001) as an independent indicator of DFS (Table 5). These results confirm the prognostic value of CCND2 methylation levels for hepatocellular carcinoma.

Kaplan-Meier survival curves used to examine the correlation between cyclin D2 (CCND2) methylation status and survival of hepatocellular carcinoma (HCC) patients after hepatectomy.
(A) Overall survival curves (log-rank test, P = 0.005).
(B) Disease-free survival curves (log-rank test, P < 0.001).

Univariate analysis of variables related to overall survival of patients with hepatocellular carcinoma.
AFP, alpha-fetoprotein; TNM, tumor node metastasis; CCND2, cyclin D2; PMR, percentage methylated reference.
*significant difference (P < 0.05).

Independent risk factors for overall survival of patients with hepatocellular carcinoma.
HR, hazard ratio; CI, confidence interval; TNM, tumor node metastasis; CCND2, cyclin D2.
*significant difference (P < 0.05).
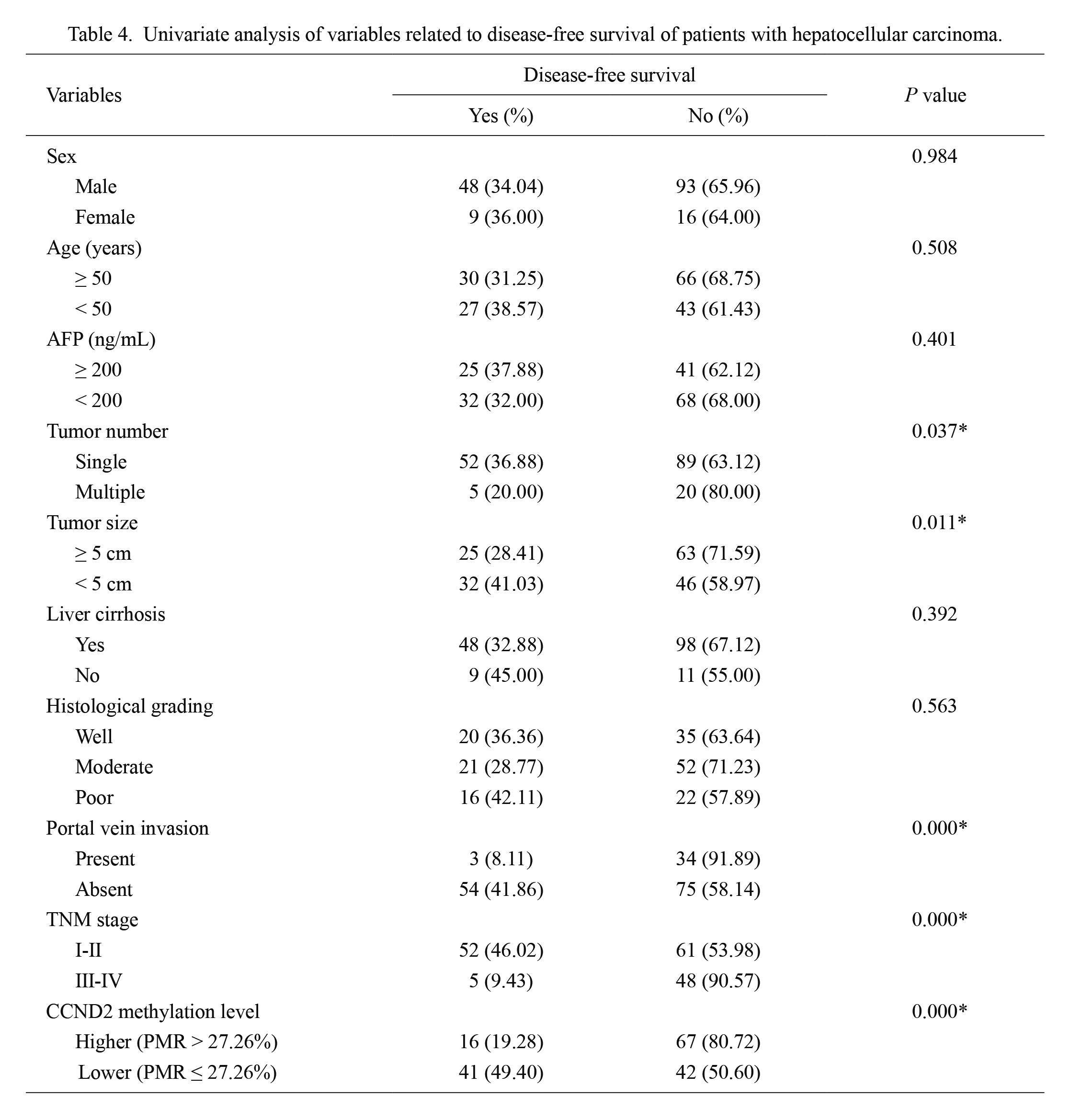
Univariate analysis of variables related to disease-free survival of patients with hepatocellular carcinoma.
AFP, alpha-fetoprotein; TNM, tumor node metastasis; CCND2, cyclin D2; PMR, percentage methylated reference.
*significant difference (P < 0.05).

Independent risk factors for disease-free survival of patients with hepatocellular carcinoma.
HR, hazard ratio; CI, confidence interval; TNM, tumor node metastasis; CCND2, cyclin D2.
*significant difference (P < 0.05).
Finally, we examined the effects of CCND2 methylation on early tumor recurrence. Among the 83 hepatocellular carcinoma patients in the lower CCND2 methylation level group, 15 (18.07%) exhibited early tumor recurrence. However, early tumor recurrence was observed in 32/83 (38.55%) of the hepatocellular carcinoma patients in the higher CCND2 methylation level group (P = 0.004), suggesting that early tumor recurrence was more common in the latter. More importantly, multivariate analyses identified higher CCND2 methylation levels (odds ratio (OR), 2.467; 95% CI, 1.143-5.324; P = 0.021) as an independent predictor of early tumor recurrence (Table 6).
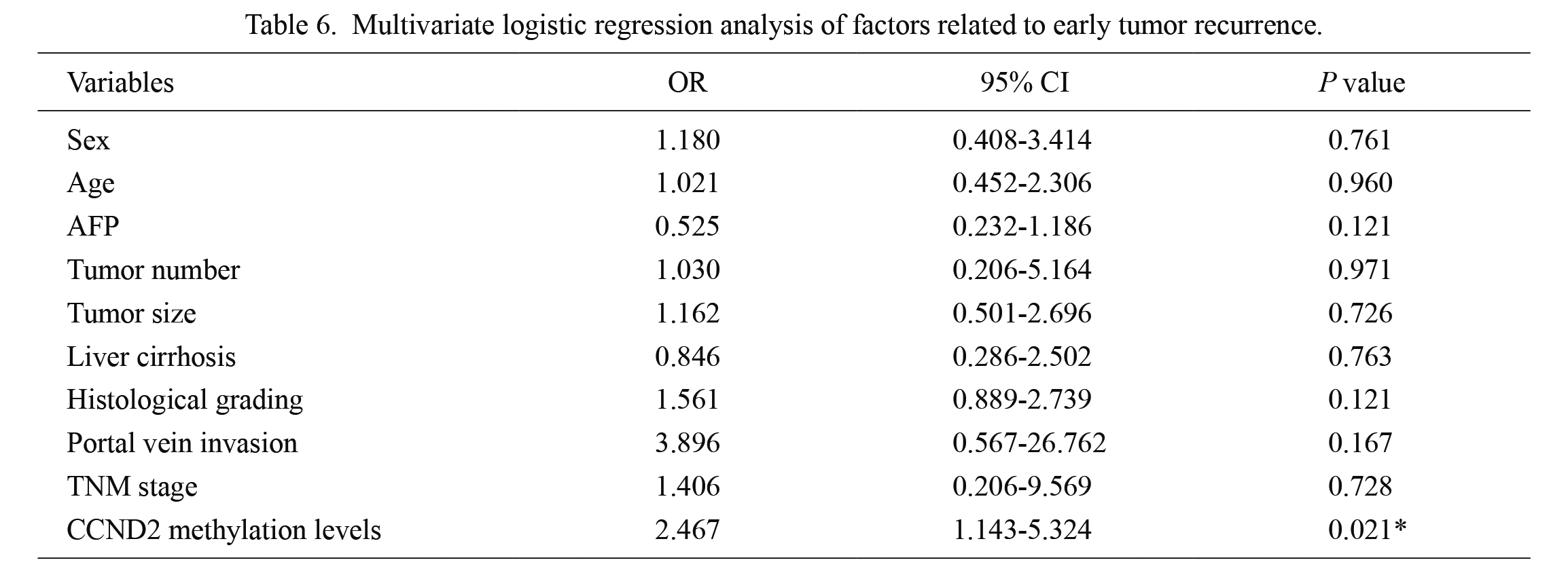
Multivariate logistic regression analysis of factors related to early tumor recurrence.
OR, odds ratio; CI, confidence interval; AFP, alpha-fetoprotein; TNM, tumor node metastasis; CCND2, cyclin D2.
*significant difference (P < 0.05).
Hepatocellular carcinoma is a common malignant tumor that progresses rapidly and has a poor prognosis. Aberrant methylation of CCND2 is associated with hepatocellular carcinoma carcinogenesis. Here, we found that the methylation levels of the CCND2 promoter in patients with HBV-associated hepatocellular carcinoma were significantly higher than that in CHB patients or healthy controls. In those with hepatocellular carcinoma, CCND2 methylation levels were higher in patients with portal vein invasion, in those with early tumor recurrence, in those with advanced TNM stage (TNM III/IV), and in those with tumor size ≥ 5 cm (P < 0.05). More importantly, hepatocellular carcinoma patients with hypermethylation of CCND2 who underwent surgical resection had poorer OS and DFS than those with lower CCND2 methylation levels. Finally, we found that CCND2 promoter hypermethylation was an independent risk factor for OS, DFS, and early tumor recurrence in hepatocellular carcinoma patients after hepatectomy.
DNA methylation is a common characteristic of tumorigenesis. Several studies show that the methylation of gene promoter regions inactivates tumor-suppressor genes and plays an important role in hepatocellular carcinoma carcinogenesis and progression (Herman and Baylin 2003; Harder et al. 2008; Yuan et al. 2016). Therefore, abnormal methylation of specific genes can be used as an early diagnosis marker and/or as an indicator of prognosis.
CCND2 regulates G1/S transition. Its main function is to form a complex with cyclin-dependent kinase (CDK)4 and CDK6; this complex induces phosphorylation of retinoblastoma (RB). Phosphorylated RB releases transcription factors, thereby activating related genes and prompting cells to enter S phase (Hunter and Pines 1994; Sherr 1995; Meyyappan et al. 1998). It is worth noting that CCND2 inhibits cell proliferation by inducing a senescence-like phenotype (Meyyappan et al. 1998), and that abnormal methylation of CCND2 plays a role in progression and poor prognosis of prostate, colorectal, and breast cancers (Evron et al. 2001; Richiardi et al. 2009; Amatu et al. 2016).
Hypermethylation of the CCND2 promoter is reported to lead to transcriptional silencing of CCND2 in many types of tumor (Evron et al. 2001; Wang et al. 2016). In addition to its role in the G1/S transition during the cell cycle, CCND2 may also function to maintain cells in the non-proliferative state and to promote exit from the cell cycle (Meyyappan et al. 1998; Mehrotra et al. 2004). A recent study revealed that knockdown of CCND2 accelerates lung cancer cell proliferation and metastasis, suggesting that CCND2 can inhibit cancer cell growth and migration (Hung et al. 2018). Chen et al. (2017) reported that CCND2 knockdown promotes cell proliferation and that restoration of CCND2 expression inhibits cell growth of prostate cancer, indicating that CCND2 may act as a tumor progression suppressor in prostate cancer. In our study, a negative correlation between CCND2 methylation and CCND2 expression in hepatocellular carcinoma patients was observed. On the basis of the above data and our results, we believe that reduced CCND2 expression may promote hepatocellular carcinoma progression. Furthermore, we found that hepatocellular carcinoma patients with portal vein invasion, early tumor recurrence, or TNM III/IV stage had high CCND2 methylation levels (Fig. 4), indicating that CCND2 methylation might be involved in tumor progression. We also found that hepatocellular carcinoma patients with higher CCND2 promoter methylation levels had a significantly shorter OS and DFS than patients with lower methylation levels (P = 0.005 and P < 0.001 respectively) (Fig. 5). Moreover, multivariate analyses confirmed the important role of CCND2 methylation levels for predicting OS and DFS (Tables 3 and 5). In accordance with our findings, Hung et al. (2018) reported that CCND2 is involved in progression and prognosis of lung cancer and breast cancer, and that hypermethylation of CCND2 correlates strongly with a poorer prognosis in lung or breast cancer patients. Our findings, along with those of the above studies, suggest that CCND2 methylation predicts a poor prognosis for patients with HBV-associated hepatocellular carcinoma after surgical resection. It is noteworthy that high CCND2 methylation levels correlated with portal vein invasion (Fig. 4A-D). We speculate that higher methylation levels of the CCND2 promoter in hepatocellular carcinoma patients with portal vein invasion may downregulate CCND2 expression, which in turn, may alleviate CCND2-mediated inhibition of cell migration (Chen et al. 2017; Hung et al. 2018). However, further study will be needed to confirm the pathway and identify the underlying mechanism.
Postoperative recurrence of hepatocellular carcinoma is complicated, and is affected by many factors (Hao et al. 2017; Imamura et al. 2003). We found that the incidence of early tumor recurrence was significantly higher in the CCND2 methylated group than that in the unmethylated group (40% vs. 15.12%, respectively; P = 0.000). In addition, multivariate analysis identified CCND2 methylation as an independent predictor of early tumor recurrence of hepatocellular carcinoma. Tsutsui et al. (2010) reported that hypermethylation of CCND2 is closely connected with early intrahepatic recurrence in patients with HCV-related hepatocellular carcinoma. Chen et al. (2017) demonstrated that CCND2 shows a strong correlation with biochemical recurrence of prostate tumors, and that CCND2 may be a potential biomarker for intermediate-risk patients that may relapse (i.e., those with a Gleason score of 7). These findings confirm the critical role of CCND2 methylation in predicting early tumor recurrence and a poor prognosis for patients with HBV-related hepatocellular carcinoma. Therefore, early and frequent detection of such methylations will help to individualize treatments and aid postoperative follow-up of patients who are CCND2 methylation-positive.
One limitation of this study is that patient PBMCs were used instead of hepatocellular carcinoma tissue. This was mainly because more convenient approaches are available for studying DNA methylation profiles or expression levels of tumor genes in PBMCs than in hepatocellular carcinoma tissue and the methylation profiles obtained from such analyses can be used as biomarkers for the diagnosis and prognosis of solid tumors (Pufulete et al. 2003; Terry et al. 2011; Zhuang et al. 2020). Moreover, Udali et al. (2015) observed that the methylcytosine statuses of hepatocellular carcinoma tissue and PBMCs are very similar. A recent study reported that the DNA methylation status in PBMCs is the same as that in hepatocellular carcinoma tumors, indicating that measuring DNA methylation status in PBMCs is a clinically valuable method that can improve disease monitoring of hepatocellular carcinomas and other tumors (Sprang et al. 2020). Furthermore, previous research has indicated that CCND2 methylation levels are markedly high in hepatocellular carcinoma tissue, and that they have potential for the diagnosis and supervision of hepatocellular carcinoma (Moribe et al. 2009; Wang et al. 2012). Therefore, in this study, the CCND2 methylation status in PBMCs was considered to reflect that in hepatocellular carcinoma tumors. In future studies, we intend to examine the CCND2 methylation status in hepatocellular carcinoma tissues to further explore the role of CCND2 in hepatocellular carcinoma.
In conclusion, hypermethylation of CCND2 was associated with hepatocellular carcinoma progression and early tumor recurrence, and served as an independent predictor for OS, DFS, and early tumor recurrence of hepatocellular carcinoma, indicating that detection of CCND2 methylation may be a non-invasive approach to predicting a poor prognosis and early tumor recurrence in patients with HBV-associated hepatocellular carcinoma undergoing hepatectomy. Furthermore, it may also have clinical value for risk stratification and monitoring of disease progression.
This work was supported by the Key Project of Chinese Ministry of Science and Technology (2017ZX102022022, 2018ZX10302206); the National Natural Science Foundation of China (81970522); and Shandong University multidisciplinary research and innovation team of young scholars (2020QNQT11).
The authors declare no conflict of interest.