Abstract
MicroRNA-152 (miR-152) expression has been reported to be associated with poor prognosis in patients with endometrial serous carcinoma (ESC). However, the function of miR-152 in ESCs is not fully understood. The present study aimed to investigate the involvement of miR-152 in ESC progression. The influence of miR-152 overexpression on cell proliferation and motility was assessed by transfecting two human ESC cell lines, USPC-1 and SPAC-1-L, with a miR-152 precursor. MiR-152 overexpression increased apoptosis and inhibited the proliferation of the two ESC cell lines. Cell motility was also suppressed in both cell lines following precursor transfection. Conversely, miR-152 inhibitor transfection led to an increase in cell migration ability, suggesting the involvement of miR-152 in ESC cell motility. Results of the analysis of publicly available messenger RNA dataset indicated that high expression of matrix metalloproteinase 10 (MMP10), one of the predicted targets of miR-152 by microRNA target prediction database, was a poor prognostic factor for ESC. In vitro examination results revealed that miR-152 overexpression reduced MMP10 expression, and knockdown of MMP10 significantly reduced cell motility. This study elucidates the function of miR-152 as a tumor suppressor in ESCs. We demonstrated that miR-152 plays an important role in ESC cell motility by regulating MMP10 expression.
Introduction
Endometrial serous carcinoma (ESC) is an aggressive subtype of endometrial carcinoma. In addition to endometrial clear cell carcinoma, ESC is classified as a type II endometrial cancer (Bokhman 1983; Hecht and Mutter 2006). In general, ESC is highly prevalent among elderly women, has less association with estrogen exposure, and shows a more progressive phenotype than type I endometrial cancers represented by grade 1 or grade 2 endometrioid carcinoma (Bokhman 1983; Sherman 2000; Hecht and Mutter 2006; Shigeta et al. 2017). Recent molecular analyses have also identified distinct genomic alterations unique to ESC or serous-like endometrial cancer (Levine et al. 2013). It is known that ESC is markedly distinct from type I endometrial carcinoma. However, the therapeutic strategies for endometrial cancer have not yet been tailored according to subtype.
MicroRNAs (miRNAs) are small non-coding RNAs that bind to messenger RNAs (mRNAs) in a sequence-dependent manner, resulting in target mRNA degradation (Bartel 2004). In addition, miRNA function is essential for maintaining normal biological processes, and miRNA expression is frequently dysregulated in cancer by several mechanisms, such as copy number alteration, aberrant transcriptional control, and dysregulated epigenetic changes (Peng and Croce 2016). Accumulating evidence has suggested that dysregulated miRNA expression is responsible for cancer development and progression (Hayes et al. 2014; Peng and Croce 2016).
Our previous studies have demonstrated that microRNA-34b (miR-34b) functions as a tumor suppressor, and let-7c contributes to paclitaxel resistance in ESC cells (Hiroki et al. 2012; Sato et al. 2020). In a previous study, we also reported reduced miR-152 and miR-101 levels as independent factors of poor prognosis for patients with ESC by analyzing surgically removed specimens (Hiroki et al. 2010). Among these, the expression of miR-101 was associated with the aggressive progression of type II endometrial cancer cell lines, including the ESC cell line SPAC-1-L (Konno et al. 2014). MiR-152 has also been identified as a tumor suppressor miRNA in endometrial cancer (Tsuruta et al. 2011). However, the detailed biological function of miR-152 in ESCs has not been investigated to the same extent as miR-101. The present study investigated the involvement of miR-152 in ESCs by focusing on cell proliferation and motility. The clinical relevance between the predicted targets of miR-152 and patient outcome was also examined by analyzing the target prediction database and the Cancer Genome Atlas (TCGA) dataset.
Methods
Endometrial carcinoma cell lines
A total of four ESC cell lines; UPSC-1, USPC-2, SPAC-1-L, SPAC-1-S were used in the current study. USPC-1 and USPC-2 cells were kindly provided by Dr. Santin, Department of Obstetrics and Gynecology, Division of Gynecologic Oncology at the Yale University School of Medicine (New Haven, CT, USA) (Santin et al. 2002). SPAC-1-L and SPAC-1-S cells were kindly provided by Dr. Hirai, Department of Gynecology, Cancer Institute Hospital of Japanese Foundation for Cancer Research (Tokyo, Japan) (Hirai et al. 1994). Ishikawa 3-H-12 and HEC-1A cells were purchased from the Japan Collection of Research Bioresources Cell Bank (Osaka, Japan). Sawano and RL95-2 cells were purchased from the RIKEN BioResource Center (Tsukuba, Japan) and the American Type Culture Collection (Manassas, VA, USA), respectively. Ishikawa 3-H-12, HEC-1A and RL95-2 cells are reported as type I endometrial cancer cell lines (Van Nyen et al. 2018). Sawano cells are reported as a cell line without estrogen receptor expression derived from moderately differentiated endometrial adenocarcinoma (Satoh et al. 1995; Fan et al. 2000). USPC-1, USPC-2, SPAC-1-L SPCA-1-S, and HEC-1A cells were cultured in Roswell Park Memorial Institute 1640 medium supplemented with 10% fetal bovine serum (FBS), 1% GlutaMAX™ (Thermo Fisher Scientific, Waltham, MA, USA), and 1% antibiotic-antimycotic (Thermo Fisher Scientific). Ishikawa 3-H-12, Sawano, and RL95-2 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS and 1% MEM-non-essential amino acids solution (Thermo Fisher Scientific).
Transfection of miRNA precursors or miR-152 inhibitor
Ambion™ Pre-miR™ miRNA Precursor Molecule for miR-152-3p (miR-152 precursor, PM12269), miR-101-3p (miR-101 precursor, PM11414), and Pre-miR™ miRNA Precursor Negative Control for miRNA precursors (miR-NC) were purchased from Thermo Fisher Scientific. A final concentration of 41.6 nM of each precursor was transfected into cells using Lipofectamine™ RNAiMAX (Thermo Fisher Scientific) following the manufacturer’s protocol. The AccuTarget™ Human miRNA Inhibitor for miR-152 and the negative control (SMC-2101) were provided by Bioneer Corporation (Daejeon, Korea). MiR-152 inhibitor (20 nM) or negative control was transfected into cells using Lipofectamine™ RNAiMAX.
Quantitative polymerase chain reaction (PCR) assays
For mRNA quantification, RNA was extracted from cells using ISOGEN II (Nippon Gene, Tokyo, Japan) and reverse-transcribed to complementary DNA using ReverTra Ace qPCR RT Master Mix and gDNA Remover (TOYOBO, Osaka, Japan). For miRNA quantification, RNA was extracted using the miRNeasy Mini Kit (Qiagen, Venlo, Netherlands) and reverse-transcribed with the Taqman MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific). Quantitative PCR was performed using TaqMan™ Fast Advanced Master Mix (Thermo Fisher Scientific) with a pre-designed TaqMan probe and primer sets targeting DNMT1 (Hs00945875_m1), EZH2 (Hs00544830_m1), matrix metalloproteinase (MMP) 10 (Hs00233987_m1), MMP3 (Hs00968305_m1), MMP13 (Hs00942584_m1), MMP15 (Hs00233997_m1), MMP19 (Hs00419424_m1), GAPDH (4333764T), miR-152 (000475), and RNU6B (001093) (Thermo Fisher Scientific) according to the manufacturer’s instructions. Relative mRNA expression and miRNA expression were determined with respect to GAPDH expression and RNU6B expression, respectively.
Cell viability and caspase 3/7 assays
Cell viability was assessed with a water-soluble tetrazolium salt assay using the Cell Counting Kit-8 (Dojindo, Kumamoto, Japan). Relative caspase 3/7 activity was measured 48 h after the transfection of miRNA precursors using a Caspase-Glo 3/7 Assay kit purchased from Promega (Madison, WI, USA).
Dicer-substrate small interference RNA (dsiRNA) transfection
Two independent dsiRNAs targeting MMP10 and a negative control dsiRNA were purchased from Integrated DNA Technologies (Coralville, IA, USA). Each dsiRNA (10 nM) was transfected into USPC-1 and SPAC-1-L cells using Lipofectamine™ RNAiMAX according to the manufacturer’s instructions.
Transwell assay
Cell migration was assessed using a Transwell migration assay. 2.5 × 104 of USPC-1 cells or 5.0 × 104 of SPAC-1-L cells suspended in a medium supplemented with 1% bovine serum albumin were added to a cell culture insert (353097) purchased from Corning Inc. (Corning, NY, USA). Each insert was then placed in a well of a 24-well plate filled with the culture medium. After incubation at 37°C for 6 h and removal of non-migrated cells, the migrated cells were fixed with methanol and stained with toluidine blue. The total number of cells was counted manually using a phase contrast light microscope by selecting five random fields of view in each well at 100× magnification.
Western blotting
Primary antibodies against MMP10 (113496) and β-actin (NB600-501) were obtained from Genetex (Irvine, CA, USA) and Novus Biologicals (Centennial, CO, USA), respectively. Mouse horseradish peroxidase-linked secondary antibodies were purchased from Cytiva (NA931; Tokyo, Japan). Rabbit HRP-linked secondary antibodies (7074S) and anti-PARP antibodies (9542) were purchased from Cell Signaling Technology Japan (Tokyo, Japan). Proteins were extracted using M-PER™ Mammalian Protein Extraction Reagent (Thermo Fisher Scientific). Thereafter, an equal amount of total protein was loaded into each well, subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis, and transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 5% dry fat milk in Tris-buffered saline with 0.05% Tween-20 (TBS-T) and incubated with primary antibodies diluted at ratios of 1:500 (anti-MMP10), 1:1,000 (anti-PARP), or 1:5,000 (anti-β-actin) in Can Get Signal Solution (TOYOBO) at 4°C overnight. After washing the membrane with TBS-T, the membrane was incubated with secondary antibodies diluted at a ratio of 1:10,000 in Can Get Signal Solution for 1 h at room temperature. Blots were developed and detected using ECL Prime Western Blotting Detection Reagent (Cytiva) and the VersaDoc Imaging System (Bio-Rad Laboratories, Hercules, CA, USA).
TCGA data analysis
The mRNA expression z-scores of MMP3, MMP10, MMP13, MMP15, and MMP19 relative to all samples and survival information recorded in TCGA were obtained using cBioPortal (Cerami et al. 2012; Gao et al. 2013). In total, 529 cases, recorded as endometrial cancer in PanCancer Atlas by TCGA, were assessed (Hoadley et al. 2018). Three patients with missing survival information or mRNA expression data were excluded. Survival analysis was performed using the Kaplan-Meier method (Kaplan and Meier 1958). The difference in overall survival between the two groups was compared using the log-rank test (Peto and Peto 1972).
Statistical analysis
Unless otherwise mentioned, the results are presented as the mean ± standard deviation of three or four independent experiments and Student’s t-test was used to analyze statistical differences. Statistical significance was set at P < 0.05. Statistical analyses were performed using GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA).
Results
MiRNA precursor transfection effectively and specifically regulates the expression of known target genes
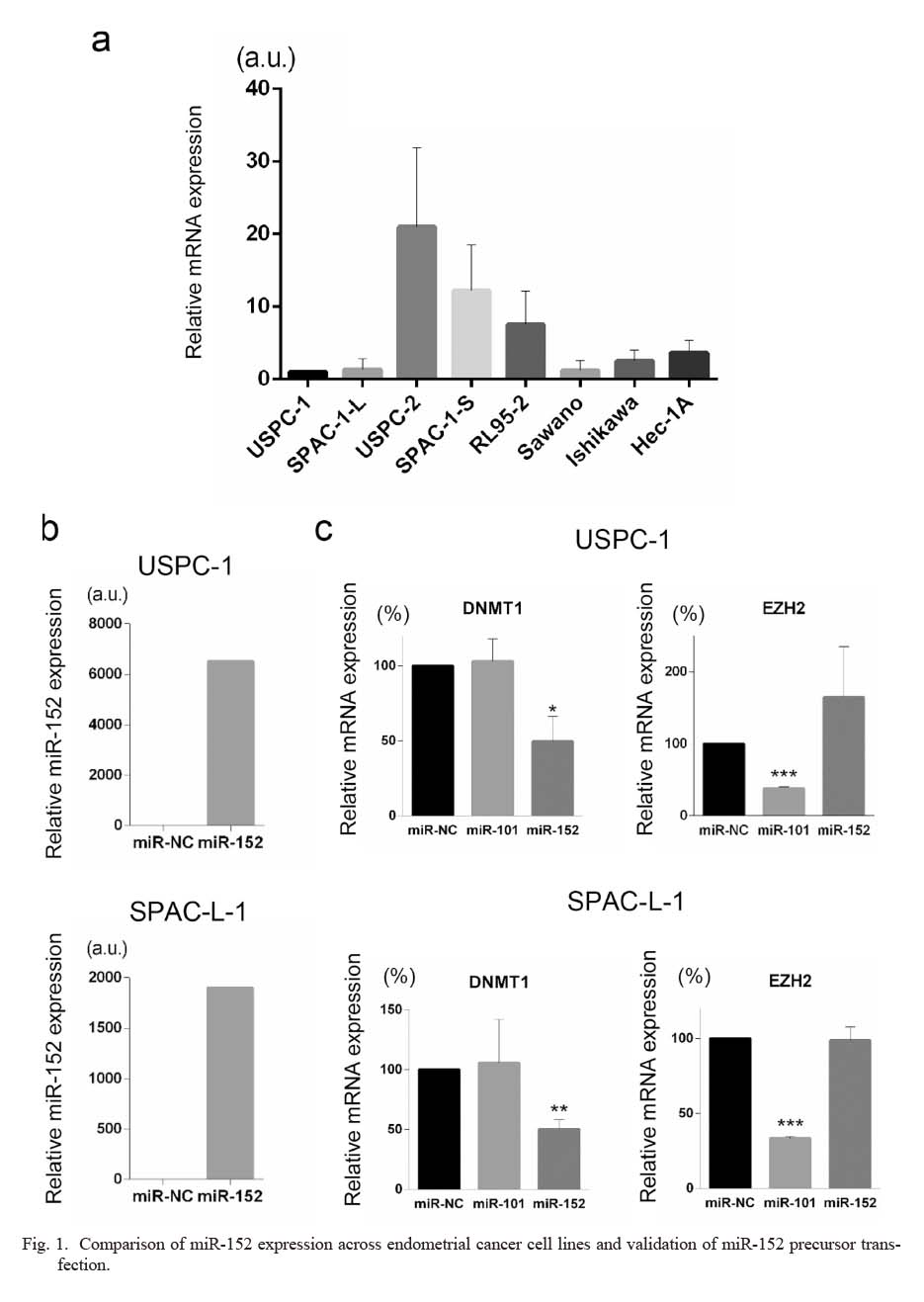
We first compared miR-152 expression among several endometrial cancer cell lines, including four ESC cells; USPC-1, USPC-2, SPAC-1-L, and SPAC-1-S. As shown in Fig. 1a, miR-152 expression varied among ESC cell lines. USPC-1 and SPAC-1-L cells, which exhibit lower miR-152 expression than other ESC cells, were selected for subsequent miR-152 overexpression assays with precursor transfection. qPCR assay results revealed successful overexpression of miR-152 in both cell lines (Fig. 1b). To further validate the efficacy and specificity of the miR-152 precursors used in this study, we examined the expression of DNMT1 and EZH2 after miRNA precursor transfection. DNMT1 is a well-established target of miR-152 (Braconi et al. 2010; Tsuruta et al. 2011; Xiang et al. 2014). EZH2 is a known target of miR-101 but not miR-152 (Friedman et al. 2009; Konno et al. 2014; Wang et al. 2016). The influence of miR-152 or miR-101 precursor transfection on DNMT1 and EZH2 expression was examined by relative quantification of mRNA. As shown in Fig. 1c, miR-152 precursor transfection reduced DNMT1 mRNA expression but did not significantly affect EZH2 expression. In contrast, miR-101 precursor transfection significantly reduced the expression of EZH2 but not that of DNMT1. These results validate the efficacy and specificity of the miR-152 precursor used in this study and its efficient transfection in USPC-1 and SPAC-1-L cell lines.
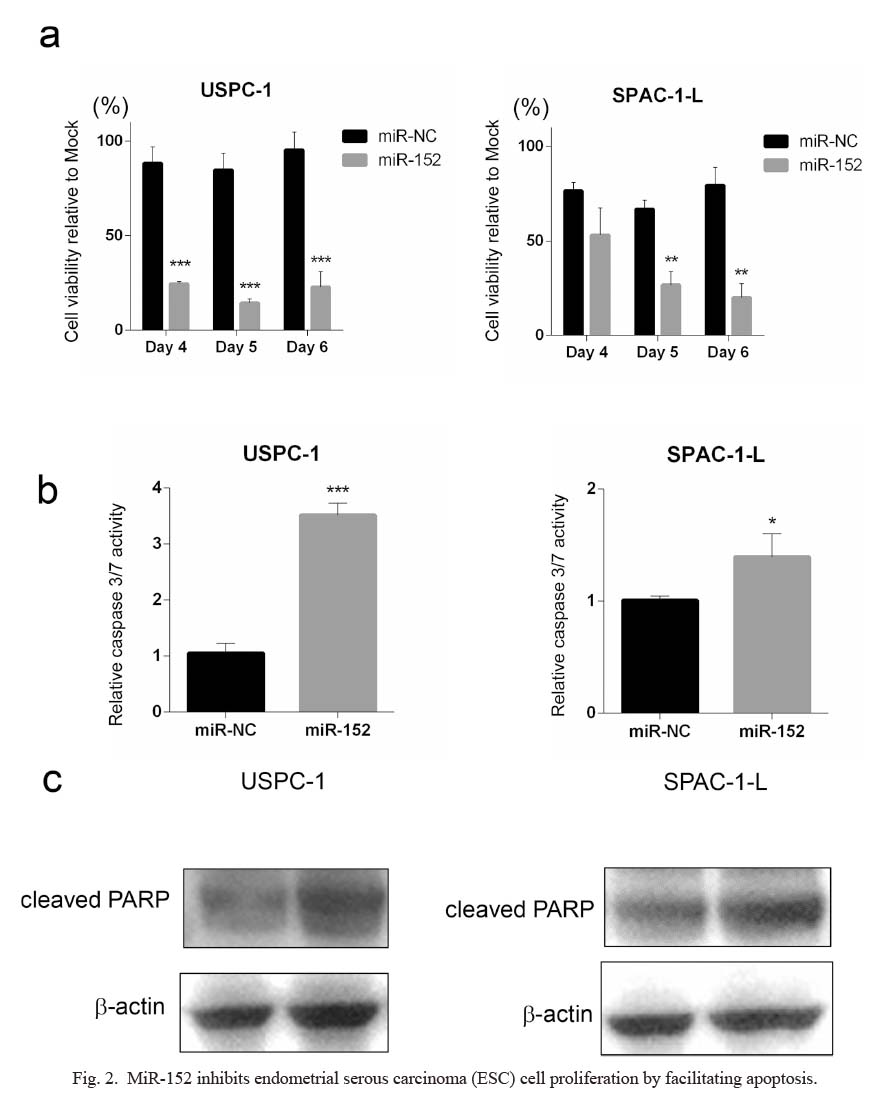
To understand the mechanism through which miR-152 overexpression affects the ESC phenotype, the miR-152 precursor was transfected into ESC cells, and cell viability was examined. Compared with miR-NC transfection, miR-152 transfection significantly suppressed tumor proliferation in both USPC-1 and SPAC-1-L cells (Fig. 2a).
Since miR-152 has been reported to mediate apoptosis in tumor cells (Sun et al. 2017; Yang et al. 2018), we further evaluated whether miR-152 is involved in apoptosis in ESCs by determining caspase 3/7 activity. In USPC-1 and SPAC-1-L cells, miR-152 precursor transfection increased caspase 3/7 activity compared to control (Fig. 2b). If there were no marked changes, the levels of cleaved form of PARP mildly increased with miR-152 precursor transfection, consistent with the increased caspase 3/7 activity (Fig. 2c). The results in Fig. 2 suggest that miR-152 suppresses ESC cell proliferation in an apoptosis-mediated manner.
MiR-152 overexpression inhibits cell migration and downregulates of MMP10 expression
Similar to cell proliferation, cancer cell motility is an important factor that regulates cancer progression. Thereafter, we next evaluated the involvement of miR-152 in ESC cell migration. As shown in Fig. 3a, miR-152 precursor transfection significantly inhibited the migration of USPC-1 and SPAC-1-L cells. Conversely, transfection with miR-152 inhibitor significantly decreased the expression of miR-152 and facilitated USPC-1 cell migration (Fig. 3b, c). Collectively, the results shown in Fig. 3 validate the involvement of miR-152 in ESC cell motility.
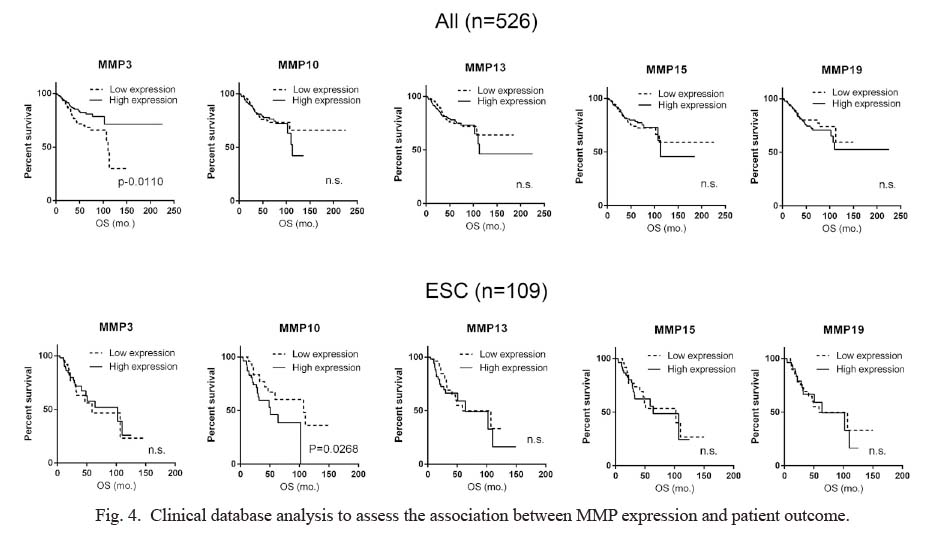
To identify candidate targets of miR-152 that may be responsible for regulating cell motility, we used the miRNA target prediction database (miRDB) (Liu and Wang 2019; Chen and Wang 2020). MiRDB predicted 838 candidate genes as possible targets of miR-152-3p. Among the candidates, we focused on the MMP family because it has been widely documented to be involved in cancer cell motility, and MMP3 is a validated target of miR-152 in glioma cells, which led us to hypothesize that other MMPs might also be regulated by miR-152 (Coussens et al. 2002; Nabeshima et al. 2002; Vihinen and Kähäri 2002; Zheng et al. 2013; Cathcart et al. 2015). Although MMP3 was not involved in the predicted targets, four MMPs were found among the 838 predicted targets: MMP10, MMP13, MMP15, and MMP19. To investigate the significance of MMP3 and these MMPs, the correlation between the expression of each MMP at the mRNA level and OS of the patients was analyzed in a clinical setting, using the data of 526 patients with endometrial cancer, including 109 ESC patients, registered in PanCancer-Atlas. For each MMP, the cases were sorted by relative mRNA expression z-score and divided into high expression (above the 50th percentile) and low expression (at or below the 50th percentile) groups. OS between the two groups was compared to determine statistical significance. Interestingly, the patients presenting higher MMP3 expression showed significantly better OS than those with lower MMP3 expression in all endometrial cancer cases, which was contrary to our expectations. There was no significant difference observed between the two groups when limited to patients with ESC. In contrast, higher MMP10 expression was significantly associated with worse OS in patients with ESC but not in patients with all types of endometrial cancer. For other MMPs, no significant differences were observed in either ESC or all endometrial cancer cases (Fig. 4).
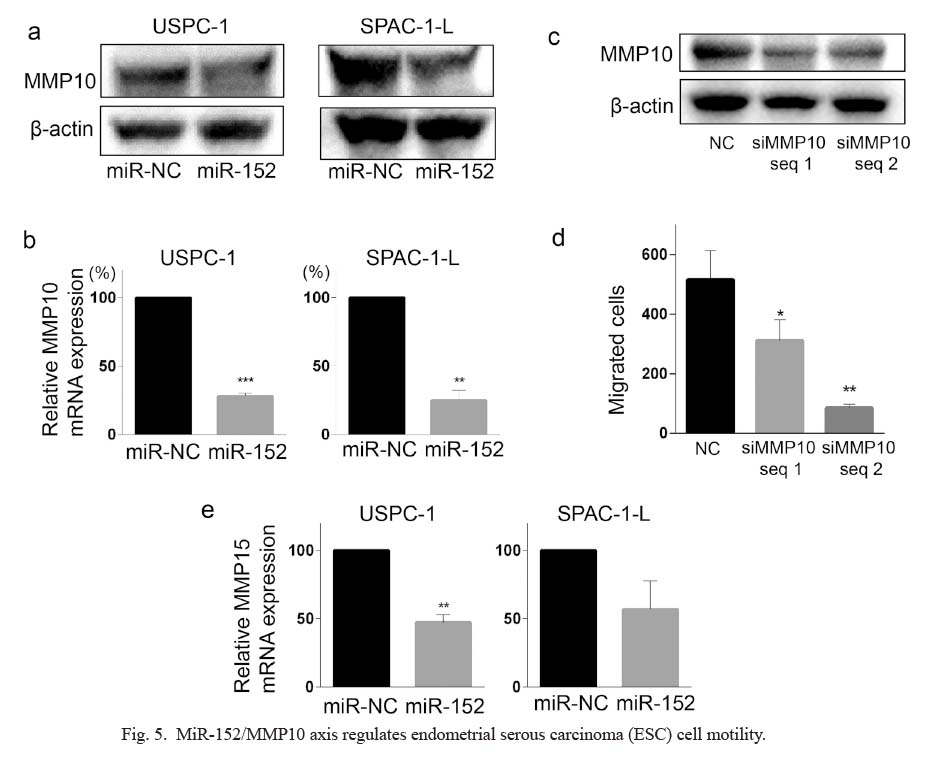
Based on this result, we examined the effect of miR-152 precursor transfection on MMP10 protein expression. MiR-152 precursor transfection reduced the expression of MMP10 in both USPC-1 and SPAC-L-1 cells, as shown in Fig. 5a. Quantitative PCR validated that miR-152 precursor transfection significantly decreased the relative expression of MMP10 mRNA, indicating MMP10 as a direct target of miR-152 (Fig. 5b). Notably, MMP10 knockdown by dsiRNA transfection significantly decreased the motility of USPC-1 cells (Fig. 5c, d). Collectively, these data indicate that miR-152 plays a role in ESC cell motility by regulating MMP10 expression. To further address whether miR-152 regulates the other MMPs expression analyzed in Fig. 4, the influence of miR-152 precursor transfection on the expression of MMP3, MMP13, MMP15, and MMP19 was also examined in USPC-1 and SPAC-1-L cells. As shown in Fig. 5e, MMP15 expression was significantly decreased by miR-152 precursor transfection in USPC-1 cells. Although not statistically significant, MMP 15 expression also tended to be decreased in SPAC-1-L cells. On the other hand, it was difficult to appropriately assess the involvement of miR-152 in the expression of MMP3, MMP13, and MMP19 by quantitative PCR since cycle threshold values for these MMPs were less than 35 in both USPC-1 and SPAC-1-L cells, which implies that the expression of MMP3, MMP13, or MMP19 is quite low in the ESC cells.

Discussion
The present study clarified the function of miR-152 as a tumor-suppressive miRNA in ESCs. While MMPs are the targets of a various miRNAs (Li and Li 2013; Abba et al. 2014), the interaction of miR-152 with MMP expression has not been completely elucidated, except for a study by Zheng et al. (2013), which reported MMP3 as a direct target of miR-152 in glioma cells. To the best of our knowledge, this is the first study to experimentally demonstrate the involvement of miR-152 in MMP10 expression in cancer cells. MMP15 is also a possible target of miR-152 as the transfection of miR-152 precursor decreased MMP15 expression. MMP10, also known as stromelysin-2, has been reported to regulate cell motility in cervical, and head and neck cancers, consistent with our findings (Deraz et al. 2011; Zhang et al. 2014). In addition to the analysis of clinical data, our experimental results indicate that miR-152-mediated regulation of MMP10 expression may play a role in ESC cell motility and progression. Although we did not experimentally assess in the current study, it is theoretically possible that miR-152 regulates MMP10 expression not only in ESC but in the other types of endometrial carcinomas. At the same time, we estimate miR-152/MMP10 axis has a more important role in ESC with regard to caner progression when considering the result shown in Fig. 4.
We did not fully assess all miR-152 target genes directly related to cell proliferation; however, several targets of miR-152 have already been reported to be associated with cancer progression, such as DNA methyltransferase 1 (DNMT1), PI3K/AKTl (Ge et al. 2017), KLF4 (Ma et al. 2014), KLF5 (Zhang et al. 2019), and β-catenin (Wen et al. 2017). Among these, DNMT1 is one of the most widely-studied targets of miR-152. DNMT1 functions as a DNA methyltransferase, and aberrant DNMT1 expression disrupts the expression of major tumor suppressor genes (Zhang et al. 2020). DNMT1 also activates important signaling pathways for cancer proliferation, such as the PI3K/AKT and IL6/STAT3 pathways (Zhang et al. 2020). As the expression of DNMT1 was significantly reduced following the transfection of USPC-1 and SPAC-1-L cells with the miR-152 precursor, we suggest that the downregulation of miR-152 expression in ESCs disrupts DNMT1 expression, resulting in ESC cell proliferation.
MMPs appear to be promising therapeutic targets for cancer treatment. However, MMP inhibitors reported in the 2000s failed to provide a survival benefit; musculoskeletal syndrome was observed as a common adverse effect of these inhibitors (Winer et al. 2018). This suggests that the development of selective inhibitors to each MMP and the appropriate selection of the target patient population may be necessary (Cathcart et al. 2015; Winer et al. 2018). Razai et al. (2020) reported a single-domain antibody that selectively inhibits MMP10. Although we are still at the preclinical phase, the concept of selective MMP10 inhibition for ESC treatment may be beneficial from the perspective of precision medicine, and miR-152 expression status has the potential to serve as a biomarker with high sensitivity.
Unexpectedly, higher MMP3 expression was associated with significantly better overall survival in the patients with endometrial cancer in the clinical data set analysis. The result indicates MMPs are involved not only in cancer cell motility but also in various biological functions of cancer cells. We think there is room for further investigation regarding the role of MMPs in cancer biology including MMP10.
In conclusion, this study highlighted the function of miR-152 as a tumor suppressor in ESCs. We believe that by understanding transcriptional regulation by miR-152 comprehensively, it will be possible to develop novel targeted therapies for patients with ESC.
Acknowledgments
This study was financially supported by a JSPS KAKENHI grant (19H03795). This work was also supported in part by the National Cancer Center Research and Development Fund (2020-J-3). The authors would like to thank Drs. A.D. Santin and Y. Hirai for kindly providing the cell lines, Ms. E. Oba and N. Hashimoto for providing experimental support, and Editage (https://www.editage.com) for English language editing.
Conflict of Interest
The authors declare no conflict of interest.
References
-
Abba,
M.,
Patil,
N. &
Allgayer,
H.
(2014) MicroRNAs in the regulation of MMPs and metastasis. Cancers (Basel), 6, 625-645.
-
Bartel,
D.P.
(2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281-297.
-
Bokhman,
J.V.
(1983) Two pathogenetic types of endometrial carcinoma. Gynecol. Oncol., 15, 10-17.
-
Braconi,
C.,
Huang,
N. &
Patel,
T.
(2010) MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology, 51, 881-890.
-
Cathcart,
J.,
Pulkoski-Gross,
A. &
Cao,
J.
(2015) Targeting matrix metalloproteinases in cancer: bringing new life to old ideas. Genes Dis., 2, 26-34.
-
Cerami,
E.,
Gao,
J.,
Dogrusoz,
U.,
Gross,
B.E.,
Sumer,
S.O.,
Aksoy,
B.A.,
Jacobsen,
A.,
Byrne,
C.J.,
Heuer,
M.L.,
Larsson,
E.,
Antipin,
Y.,
Reva,
B.,
Goldberg,
A.P.,
Sander,
C. &
Schultz,
N.
(2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov., 2, 401-404.
-
Chen,
Y. &
Wang,
X.
(2020) miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res., 48, D127-D131.
-
Coussens,
L.M.,
Fingleton,
B. &
Matrisian,
L.M.
(2002) Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science, 295, 2387-2392.
-
Deraz,
E.M.,
Kudo,
Y.,
Yoshida,
M.,
Obayashi,
M.,
Tsunematsu,
T.,
Tani,
H.,
Siriwardena,
S.B.S.M, Keikhaee, M.R.,
Qi,
G.,
Iizuka,
S.,
Ogawa,
I.,
Campisi,
G.,
Lo Muzio,
L.,
Abiko,
Y.,
Kikuchi,
A.,
et al.(2011) MMP-10/stromelysin-2 promotes invasion of head and neck cancer. PLoS One, 6, e25438.
-
Fan,
H.,
Morioka,
T. &
Ito,
E.
(2000) Induction of apoptosis and growth inhibition of cultured human endometrial adenocarcinoma cells (Sawano) by an antitumor lipoprotein fraction of rice bran. Gynecol. Oncol., 76, 170-175.
-
Friedman,
J.M.,
Liang,
G.,
Liu,
C.C.,
Wolff,
E.M.,
Tsai,
Y.C.,
Ye,
W.,
Zhou,
X. &
Jones,
P.A.
(2009) The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res., 69, 2623-2629.
-
Gao,
J.,
Aksoy,
B.A.,
Dogrusoz,
U.,
Dresdner,
G.,
Gross,
B.,
Sumer,
S.O.,
Sun,
Y.,
Jacobsen,
A.,
Sinha,
R.,
Larsson,
E.,
Cerami,
E.,
Sander,
C. &
Schultz,
N.
(2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal., 6, pl1.
-
Ge,
S.,
Wang,
D.,
Kong,
Q.,
Gao,
W. &
Sun,
J.
(2017) Function of miR-152 as a tumor suppressor in human breast cancer by targeting PIK3CA. Oncol. Res., 25, 1363-1371.
-
Hayes,
J.,
Peruzzi,
P.P. &
Lawler,
S.
(2014) MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med., 20, 460-469.
-
Hecht,
J.L. &
Mutter,
G.L.
(2006) Molecular and pathologic aspects of endometrial carcinogenesis. J. Clin. Oncol., 24, 4783-4791.
-
Hirai,
Y.,
Kawaguchi,
T.,
Hasumi,
K.,
Kitagawa,
T. &
Noda,
T.
(1994) Establishment and characterization of human cell lines from a serous papillary adenocarcinoma of the endometrium. Gynecol. Oncol., 54, 184-195.
-
Hiroki,
E.,
Akahira,
J.,
Suzuki,
F.,
Nagase,
S.,
Ito,
K.,
Suzuki,
T.,
Sasano,
H. &
Yaegashi,
N.
(2010) Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci., 101, 241-249.
-
Hiroki,
E.,
Suzuki,
F.,
Akahira,
J.,
Nagase,
S.,
Ito,
K.,
Sugawara,
J.,
Miki,
Y.,
Suzuki,
T.,
Sasano,
H. &
Yaegashi,
N.
(2012) MicroRNA-34b functions as a potential tumor suppressor in endometrial serous adenocarcinoma. Int. J. Cancer, 131, E395-404.
-
Hoadley,
K.A.,
Yau,
C.,
Hinoue,
T.,
Wolf,
D.M.,
Lazar,
A.J.,
Drill,
E.,
Shen,
R.,
Taylor,
A.M.,
Cherniack,
A.D.,
Thorsson,
V.,
Akbani,
R.,
Bowlby,
R.,
Wong,
C.K.,
Wiznerowicz,
M.,
Sanchez-Vega,
F.,
et al.(2018) Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell, 173, 291-304. e296.
-
Kaplan, E. L. &
Meier,
P.
(1958) Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc., 53, 457-481.
-
Konno,
Y.,
Dong,
P.,
Xiong,
Y.,
Suzuki,
F.,
Lu,
J.,
Cai,
M.,
Watari,
H.,
Mitamura,
T.,
Hosaka,
M.,
Hanley,
S.J.,
Kudo,
M. &
Sakuragi,
N.
(2014) MicroRNA-101 targets EZH2, MCL-1 and FOS to suppress proliferation, invasion and stem cell-like phenotype of aggressive endometrial cancer cells. Oncotarget, 5, 6049-6062.
-
Levine, D. A.; The Cancer Genome Atlas Research Network
(2013) Integrated genomic characterization of endometrial carcinoma. Nature, 497, 67-73.
-
Li,
L. &
Li,
H.
(2013) Role of microRNA-mediated MMP regulation in the treatment and diagnosis of malignant tumors. Cancer Biol. Ther., 14, 796-805.
-
Liu,
W. &
Wang,
X.
(2019) Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol., 20, 18.
-
Ma,
J.,
Yao,
Y.,
Wang,
P.,
Liu,
Y.,
Zhao,
L.,
Li,
Z.,
Li,
Z. &
Xue,
Y.
(2014) MiR-152 functions as a tumor suppressor in glioblastoma stem cells by targeting Kruppel-like factor 4. Cancer Lett., 355, 85-95.
-
Nabeshima,
K.,
Inoue,
T.,
Shimao,
Y. &
Sameshima,
T.
(2002) Matrix metalloproteinases in tumor invasion: role for cell migration. Pathol. Int., 52, 255-264.
-
Peng,
Y. &
Croce,
C.M.
(2016) The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther., 1, 15004.
-
Peto,
R. &
Peto,
J.
(1972) Asymptotically efficient rank invariant test procedures. Journal of the Royal Statistical Society. Series A (General), 135, 185-207.
-
Razai,
A.S.,
Eckelman,
B.P. &
Salvesen,
G.S.
(2020) Selective inhibition of matrix metalloproteinase 10 (MMP10) with a single-domain antibody. J. Biol. Chem., 295, 2464-2472.
-
Santin,
A.D.,
Bellone,
S.,
Gokden,
M.,
Palmieri,
M.,
Dunn,
D.,
Agha,
J.,
Roman,
J.J.,
Hutchins,
L.,
Pecorelli,
S.,
O’Brien,
T.,
Cannon,
M.J. &
Parham,
G.P.
(2002) Overexpression of HER-2/neu in uterine serous papillary cancer. Clin. Cancer Res., 8, 1271-1279.
-
Sato,
I.,
Ishibashi,
M.,
Tokunaga,
H.,
Shigeta,
S.,
Sakurada,
S.,
Shimada,
M.,
Nagase,
S.,
Watanabe,
Y. &
Yaegashi,
N.
(2020) MicroRNA let-7c contributes to paclitaxel resistance via Aurora-B in endometrial serous carcinoma. Tohoku J. Exp. Med., 251, 263-272.
-
Satoh,
T.,
Nishida,
M.,
Miyazaki,
Y.,
Sugita,
M.,
Arai,
Y.,
Oki,
A.,
Kono,
K.,
Tsunoda,
H.,
Kasahara,
K. &
Kubo,
T.
(1995) Establishment of a cisplatin-resistant new human endometrial adenocarcinoma cell line, Sawano cells. Hum. Cell, 8, 67-72.
-
Sherman,
M.E.
(2000) Theories of endometrial carcinogenesis: a multidisciplinary approach. Mod. Pathol., 13, 295-308.
-
Shigeta,
S.,
Nagase,
S.,
Mikami,
M.,
Ikeda,
M.,
Shida,
M.,
Sakaguchi,
I.,
Ushioda,
N.,
Takahashi,
F.,
Yamagami,
W.,
Yaegashi,
N.,
Udagawa,
Y. &
Katabuchi,
H.
(2017) Assessing the effect of guideline introduction on clinical practice and outcome in patients with endometrial cancer in Japan: a project of the Japan Society of Gynecologic Oncology (JSGO) guideline evaluation committee. J. Gynecol. Oncol., 28, e76.
-
Sun,
J.,
Tian,
X.,
Zhang,
J.,
Huang,
Y.,
Lin,
X.,
Chen,
L. &
Zhang,
S.
(2017) Regulation of human glioma cell apoptosis and invasion by miR-152-3p through targeting DNMT1 and regulating NF2: MiR-152-3p regulate glioma cell apoptosis and invasion. J. Exp. Clin. Cancer Res., 36, 100.
-
Tsuruta,
T.,
Kozaki,
K.,
Uesugi,
A.,
Furuta,
M.,
Hirasawa,
A.,
Imoto,
I.,
Susumu,
N.,
Aoki,
D. &
Inazawa,
J.
(2011) MiR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res., 71, 6450-6462.
-
Van Nyen,
T.,
Moiola,
C.P.,
Colas,
E.,
Annibali,
D. &
Amant,
F.
(2018) Modeling endometrial cancer: past, present, and future. Int. J. Mol. Sci., 19, 2348.
-
Vihinen,
P. &
Kähäri,
V.M.
(2002) Matrix metalloproteinases in cancer: prognostic markers and therapeutic targets. Int. J. Cancer, 99, 157-166.
-
Wang,
H.,
Meng,
Y.,
Cui,
Q.,
Qin,
F.,
Yang,
H.,
Chen,
Y.,
Cheng,
Y.,
Shi,
J. &
Guo,
Y.
(2016) MiR-101 targets the EZH2/Wnt/beta-Catenin the pathway to promote the osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. Sci. Rep., 6, 36988.
-
Wen,
Y.Y.,
Liu,
W.T.,
Sun,
H.R.,
Ge,
X.,
Shi,
Z.M.,
Wang,
M.,
Li,
W.,
Zhang,
J.Y.,
Liu,
L.Z. &
Jiang,
B.H.
(2017) IGF-1-mediated PKM2/beta-catenin/miR-152 regulatory circuit in breast cancer. Sci. Rep., 7, 15897.
-
Winer,
A.,
Adams,
S. &
Mignatti,
P.
(2018) Matrix metalloproteinase inhibitors in cancer therapy: turning past failures into future successes. Mol. Cancer Ther., 17, 1147-1155.
-
Xiang,
Y.,
Ma,
N.,
Wang,
D.,
Zhang,
Y.,
Zhou,
J.,
Wu,
G.,
Zhao,
R.,
Huang,
H.,
Wang,
X.,
Qiao,
Y.,
Li,
F.,
Han,
D.,
Wang,
L.,
Zhang,
G. &
Gao,
X.
(2014) MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene, 33, 378-386.
-
Yang,
Y.,
Fang,
X.,
Yang,
R.,
Yu,
H.,
Jiang,
P.,
Sun,
B. &
Zhao,
Z.
(2018) MiR-152 regulates apoptosis and triglyceride production in MECs via targeting ACAA2 and HSD17B12 genes. Sci. Rep., 8, 417.
-
Zhang,
G.,
Miyake,
M.,
Lawton,
A.,
Goodison,
S. &
Rosser,
C.J.
(2014) Matrix metalloproteinase-10 promotes tumor progression through regulation of angiogenic and apoptotic pathways in cervical tumors. BMC Cancer, 14, 310.
-
Zhang,
H.,
Lu,
Y.,
Wang,
S.,
Sheng,
X. &
Zhang,
S.
(2019) MicroRNA-152 acts as a tumor suppressor microRNA by inhibiting Kruppel-like factor 5 in human cervical cancer. Oncol. Res., 27, 335-340.
-
Zhang,
J.,
Yang,
C.,
Wu,
C.,
Cui,
W. &
Wang,
L.
(2020) DNA methyltransferases in cancer: biology, paradox, aberrations, and targeted therapy. Cancers (Basel), 12, 2123.
-
Zheng,
X.,
Chopp,
M.,
Lu,
Y.,
Buller,
B. &
Jiang,
F.
(2013) MiR-15b and miR-152 reduce glioma cell invasion and angiogenesis via NRP-2 and MMP-3. Cancer Lett., 329, 146-154.