2022 年 257 巻 2 号 p. 135-145
2022 年 257 巻 2 号 p. 135-145
Dysregulated expression of ubiquitin-specific protease 43 (USP43) has been recently discovered in malignancies. This study aimed to investigate the expression pattern of USP43 protein in lung squamous cell carcinoma (LUSC) and to explore its correlation with patients’ clinicopathological characteristics as well as clinical outcomes. Expression of USP43 protein was determined by immunohistochemistry staining in a retrospective cohort containing 157 LUSC cases who underwent curative surgery in our hospital. Accordingly, USP43 protein was positively correlated with tumor size, depth of invasion, and lymph node metastasis. Patients with increased USP43 expression or positive lymph nodes exhibited a poorer overall survival. In addition, cellular assays elucidated that USP43 can promote LUSC growth and invasion. Taken together, our study demonstrated that USP43 may act as a proto-oncogene, which could be a promising biomarker and therapeutic target in the survival prediction and treatment of LUSC.
Lung cancer is one of the most common human malignancies and is the leading cause of cancer-related mortality worldwide. Lung cancer is comprised of three major subtypes, including lung squamous cell carcinoma (LUSC), lung adenocarcinoma (LUAD), and large cell lung carcinoma (LCLC) (Zhang et al. 2020). Among them, LUSC is the most common subtype, which results in approximately 400,000 deaths per year (Siegel et al. 2020). LUSC is characterized with high molecular complexity and most LUSC patients were diagnosed with advanced stages (Miller et al. 2016). Therefore, although great progress has been made, treatment of LUSC remains unsatisfied considering its high recurrence rate and poor prognosis (Zhang et al. 2018b). Efficient prognostic prediction of LUSC is critical to improve the survival rate and follow-up of LUSC cases. It is urgent to identify molecular biomarkers to help predict LUSC prognosis and direct novel therapeutic development (Zhang et al. 2017; Zou et al. 2019; Yan et al. 2021; Chen et al. 2021a, 2022).
Ubiquitination and phosphorylation perhaps represent the two most important post-translational protein modifications, especially in malignancies (Tang et al. 2018; Liu et al. 2019, 2021; Wang and Wang 2021). More and more studies suggest that ubiquitin plays a critical role in carcinogenesis and cancer progression (Liu et al. 2016; Gallo et al. 2017). Ubiquitin-specific protease (USP) family is a class of enzymes with protein deubiquitinating activity, comprising the majority of protein deubiquitinating diversity within mammalian cells (Pal and Donato 2014). USPs members are also emerging as prognostic factors and therapeutic targets in many diseases, including cancers (Young et al. 2019). However, USPs may play oncogenic or anti-tumor roles in different cancers. For example, USP14 was reported as an oncogene in several cancer types (Tian et al. 2014), while USP33 act as a tumor suppressor (Li et al. 2013).
Of note, even the same USP protein can have multiple effects in tumor. For example, USP24 was reported to suppress tumor cell proliferation but can also enhance tumor metastasis (Wang et al. 2017, 2018). As a novel identified USP member, the function of USP43 is largely unknown. Its role in cancers was firstly reported by Lin et al. (2017), who showed that USP43 promotes tumorigenesis through regulating cell cycle and EMT in breast cancer. Later, USP43 was reported to promote the proliferation, migration, and invasion of colorectal cancer cells through overexpression and knockdown treatments (Ye et al. 2021). A recent RNA-sequencing study also revealed an increased USP43 in osteosarcoma cells, however, its correlation with osteosarcoma prognosis was not observed (Lavaud et al. 2021). Here we aimed to explore the expression of USP43 in LUSC tissues and test its clinical significance. Meanwhile, we investigated the tumor-related roles of USP43 in LUSC via cellular experiments.
A cohort of 157 LUSC patients was retrospectively enrolled from our hospital who underwent surgical resection from May 2009 to May 2012. None of the cases accepted preoperative chemotherapy treatment. All pathological diagnoses and staging were verified by the Department of Pathology according to the American Joint Committee on Cancer (AJCC) staging system. All the cases were treated with curative R0 resection; therefore, all resection margins were negative. Patients’ basic information such as sex and age were retrieved. In brief, our cohort was comprised of 76 female patients and 81 male patients. There were 54 cases with age younger than 65-years-old. The clinicopathological characteristics, including tumor differentiation grade, tumor site, tumor laterality, tumor size in diameter, T stage, N stage and postoperative adjuvant therapy were all included in our statistical analyses (Table 1). In detail, 66 cases showed poor pathological differentiation, 72 cases showed moderate differentiation, and the other 19 cases showed well differentiation. Among the 157 patients, 100 cases showed upper lung tumor location, while the other 57 cases showed middle or lower lung tumor location. Among them, 70 cases showed left lung tumor location, and the other 87 cases showed right lung tumor location. As for the tumor size, 75 cases were less than 5.0 cm in tumor diameter, and the other 82 cases were larger. According to the invasion depth, 52 cases were staged as T1 stage, 67 cases as stage T2, 31 cases as stage T3, and the other 7 cases as stage T4. Similarly, 113 cases were classified as N0 stage, 19 cases with N1 stage, while the other 25 cases with N2 stage. After surgical treatment, 60 cases accepted adjuvant therapy, while the other 97 cases were absent of adjuvant therapy. Besides the above patients enrolled from our medical center, we also retrieved the survival data of gastric cancer patients from TCGA database (https://www.cancer.gov/tcga).
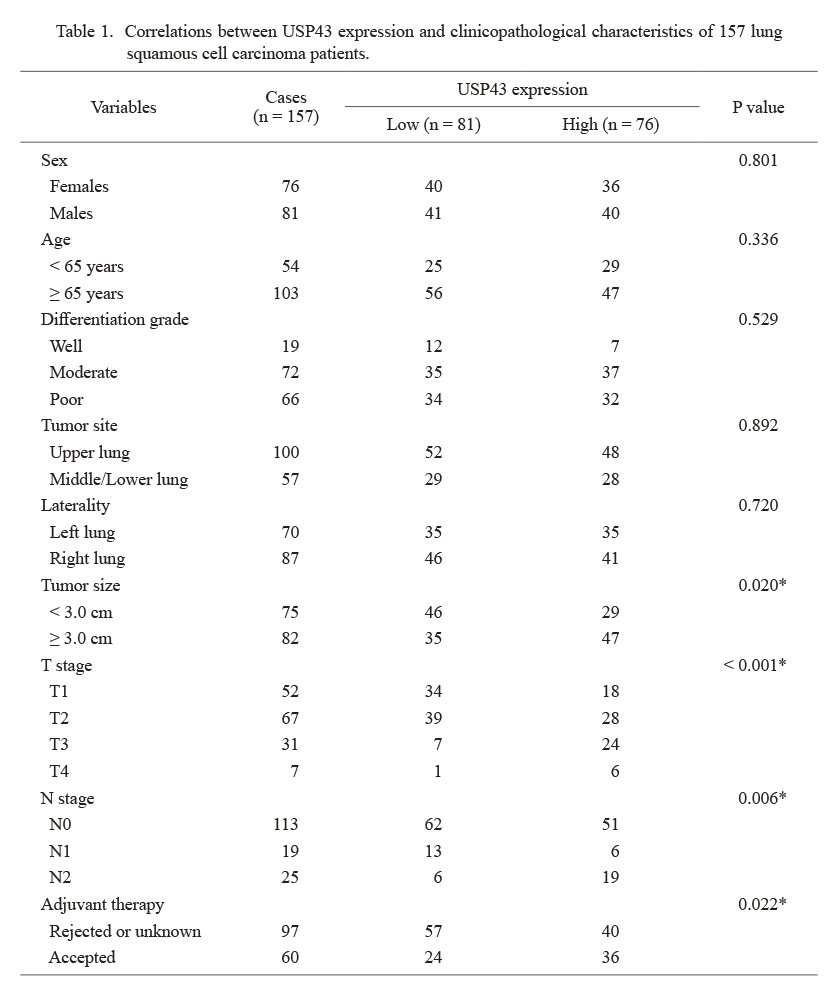
Correlations between USP43 expression and clinicopathological characteristics of 157 lung squamous cell carcinoma patients.
*Statistically significant by Chi-square test.
USP43, ubiquitin specific peptidase 43.
All the 157 paired tissue sections from formalin-fixed paraffin-embedded blocks were deparaffinized, rehydrated, and then incubated in a pH 7.8 Tris-EDTA-citrate buffer at 121°C for 20 min for antigen retrieval. Primary antibody against USP43 (1:150 dilution) (#AP14283b, Abcepta, Australia) was incubated overnight at 4°C. Then slides were incubated with secondary antibody for 30 min at room temperature (Chen et al. 2021b).
Staining results were evaluated independently by two pathologists who were blinded to patients’ characteristics. USP43 mainly localized in the cytoplasm of LUSC tissues. According to the cytoplasm staining, staining intensity was scored as negative staining (0), weak staining (1), moderate staining (2) or strong staining (3). At the same time, the percentage of stained cells was also scored: 0-25% positive cells (1), 26-50% positive cells (2), 51-75% positive cells (3), or 76-100% positive cells (4). The total immunoreactivity score was calculated by multiplication of these two scores above, ranging 0-12. According to the median staining score, patients were sub-grouped into two groups, the low-USP43 group (n = 81) and high-USP43 group (n = 76).
Cell lines and transfectionThe human LUSC cell lines (NCI-H520 and NCI-H1703) and HEK293T cells were obtained from the Cell Bank of Chinese Academy of Sciences, Shanghai Branch. All the three cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum in 5% CO2 and 90% humidity at 37°C.
The USP43 short hairpin RNA (shRNA) vector (shUSP43#1, shUSP43#2) and its negative control (scrambled shRNA) were synthesized by Genechem CO., Ltd. (Shanghai, China) as reported by others (He et al. 2018). Briefly, the recombinant constructs in pLKO.1 vectors, as well as assistant vectors psPAX2 and pMD2.G, were co-transfected into HEK293T cells. After culturing for 48 h, the viral supernatants were collected, filtered, and concentrated, and were used to infect the NCI-H520 and NCI-H1703 cells, respectively. After infection, cells were selected with puromycin (#P9620, Sigma Aldrich, St. Louis, MO, USA) at 2.0 μg/ml concentration and maintained in 0.5 μg/ml puromycin during cell culture.
Western blot analysisCultured cells were lysed in radio-immunoprecipitation assay (RIPA) buffer (containing 1% phenylmethylsulfonyl fluoride) (#G2033, Servicebio, Wuhan, China) for extraction of total protein. BCA protein Assay kit (#23225, Thermo Fisher Scientific, Pittsburg, PA, USA) was applied to quantify the protein concentration. The protein samples (20 μg) were loaded on 10% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (#1620177, Bio-Rad, Hercules, CA, USA) which were blocked in 5% non-fat milk at room temperature for 1 h. Then primary antibodies (all 1:1,000 dilutions) were used to incubate with the PVDF membranes overnight. The primary antibodies for western blot included anti-USP43 (#AP14283b, Abcepta), anti-cyclin-dependent kinase 1 (CDK1) (#ab18, Abcam, Boston, MA, USA), anti-vimentin (#sc-6260, Santa Cruz Biotechnology, Dallas, Texas, USA), anti-snail (#sc-271977, Santa Cruz Biotechnology), and anti-GAPDH (#sc-47724, Santa Cruz Biotechnology). After then, secondary antibodies conjugated with horseradish peroxidase were used to incubate the membranes for another 1 h at room temperature. Finally, protein expression levels were visualized by ECL detection system (Zhang et al. 2018a). Experiments were repeated for three independent times.
Cell proliferation assayCell viability was monitored by Cell Count Kit 8 (#CK-04, Dojindo Molecular Technologies, Tokyo, Japan) according to the manufacturer’s protocol. Briefly, infected LUSC cells were counted and seeded onto 96-well cell culture plates at a density of 2,000 cells per well. Proliferation rates were then measured by absorbance of 450 nm at 0, 1, 2, 3, and 4 days according to the standard procedure. Experiments were repeated for three independent times.
Transwell assayCell migration and invasion assays were performed with transwell chambers (#3464, Corning Biocoat, Corning, NY, USA). For the migration assay, cells were seeded into the upper chamber and incubated in FBS-free medium. The bottom chamber was filled with 20% FBS-containing medium. Following culture for 24 h, cells were fixed by paraformaldehyde and dye by crystal violet. Examination and counting of the cells were conducted under a microscope (Xu et al. 2018). The procedure of invasion assays was same to migration assays except that the chambers were pre-coated with Matrigel (0.5 mg/ml; #354234, Corning Biocoat). Experiments were repeated for three independent times.
Statistical analysesOverall survival (OS) time was defined as the months from surgery date to the date of death or the last follow-up. Patients that survived less than 2 months after surgical resection were excluded to minimize perioperative death. In our follow-up, the median survival time is 39 months after surgery, ranging from 2 to 94 months. At the end of follow-up, 44 cases died. Statistical analyses were performed using SPSS 20.0 software (SPSS Inc., Chicago, IL, USA). Data was expressed as mean ± standard deviations (SD). χ2 test was used to assess the correlations between USP43 expression and clinicopathological characteristics. Student’s t test or one-way ANOVO analysis was used to compare the differences between groups. P value less than 0.05 was considered to be statistically significant in all cases.
As shown in Fig. 1A, USP43 is predominately localized in the cytosol of LUSC cells, although it showed diverse expression levels in tissues from different patients. According to the immunoreactivity of IHC staining results, we grouped patients into low-USP43 (n = 81) and high-USP43 (n = 76) groups to assess their clinical correlations (Table 1). Chi-square tests showed that patients with larger tumor size were more prevalent to exhibit higher USP43 protein level (P = 0.020). Meanwhile, the expression level of USP43 was positively associated with patients’ T stage (P < 0.001) and N stage (P = 0.006), respectively. The positive correlations between USP43 and unfavorable characteristics indicated that USP43 may participate in LUSC progression.
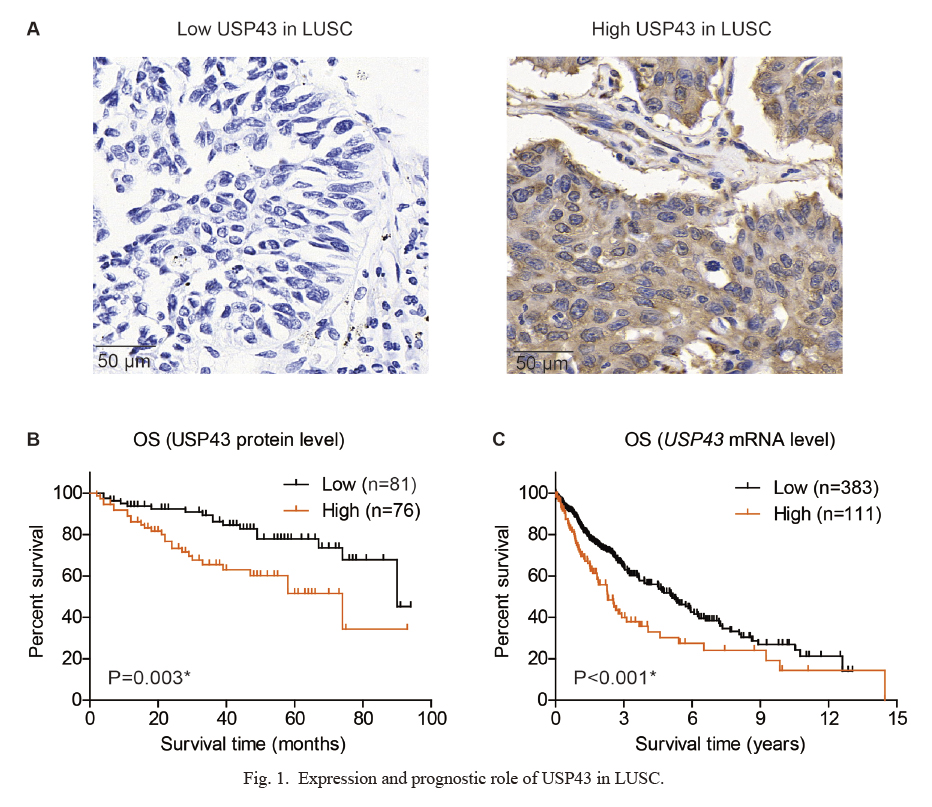
Expression and prognostic role of USP43 in LUSC.
(A) Representative high- and low-expression of USP43 protein in LUSC tissues, as reflected by the immunohistochemical staining results. (B) Kaplan-Meier survival curve of the enrolled LUSC cohort from our medical center (n = 157), showing that patients with higher USP43 protein level exhibited poorer overall survival. (C) Kaplan-Meier survival curve of TCGA cohort (n = 494), showing that patients with higher USP43-mRNA level exhibited poorer overall survival.
*P < 0.05 by log-rank test.
Kaplan-Meier method was utilized to assess the prognostic significance of USP43 protein in enrolled LUSC cohort (Fig. 1B). As a result, patients with higher USP43 showed shorter overall survival time (58.0 ± 5.1 months) compared to those with lower USP43 protein levels (76.5 ± 3.6 months). The 5-year overall survival rate of low-USP43 group was 77.8%, while it was only 51.6% of the high-USP43 group (P = 0.003). Considering that our data was obtained from a single medical center with a limited case number, we next evaluated its prognostic role using TCGA cohort (Fig. 1C). Based on its mRNA levels from microarray data, we found that patients with higher USP43 transcription level also exhibited significant poorer overall survival compared to those with lower ones (P < 0.001). Taken together, either higher level of USP43-mRNA or USP43-protein was correlated with unfavorable prognosis of LUSC patients.
Besides USP43 levels, the prognostic role of other enrolled factors was also analyzed using Kaplan-Meier method (Fig. 2, Table 2). Accordingly, poorer differentiation grade (Fig. 2A, P = 0.025), lower lung location of tumors (Fig. 2B, P = 0.022), larger tumor size (Fig. 2C, P = 0.001), advanced T stage (Fig. 2D, P = 0.034) and advanced N stage (Fig. 2E, P = 0.001) can all predict poorer overall survival of LUSC patients. Notably, patients accepted postoperative adjuvant therapies also exhibited poorer prognosis compared to those without adjuvant therapies (Fig. 2F, P < 0.001). This may be explained by the fact that only patients with advanced tumor stages were suggested for adjuvant therapy, which was significantly correlated with poorer survival.
We next conducted multivariate analysis using a Cox regression model to evaluate their independent prognostic effects. All the factors showing statistical significance (P < 0.05) by univariate analyses in Table 2 were enrolled into the Cox regression model for multivariate analysis. In detail, the confounding factors included differentiation grade, tumor site, tumor size, T stage, N stage, adjuvant therapy, and USP43 expression (Table 3). As a result, high USP43 protein level was identified as an independent unfavorable prognostic biomarker of LUSC [hazard ratio (HR) 2.165, 95% confidence interval (CI) 1.064-4.408, P = 0.033]. Meanwhile, middle or lower lung tumor location (HR 2.623, 95% CI 1.401-4.911, P = 0.003), larger tumor size (HR 2.891, 95% CI 1.142-7.319, P = 0.025), and stage N2 (HR 2.358, 95% CI 1.014-5.480, P = 0.046) all contributed independently to poorer overall survival of LUSC.
Considering the significant prediction efficiency of USP43 on LUSC survival, we further conducted stratification analyses based on T stage and N stage, respectively. As a result, high- or low-USP43 can help distinguish survival even in patients within the same T stage (Fig. 3A-C) or N stage (Fig. 3D, E). Therefore, we may conclude that monitoring USP43 expression level in LUSC tissues may help further predict patients’ outcome as a supplement to the current TNM stage, which will help direct personalized follow-up.

Overall survival curves of enrolled 157 LUSC patients.
Kaplan-Meier survival curves were generated according to the LUSC differentiation grade (A), tumor site (B), tumor size (C), T stage (D), N stage (E), or adjuvant therapy (F). *P < 0.05 by log-rank test.

Univariate analysis for the overall survival in 157 lung squamous cell carcinoma patients.
*Statistically significant by log-rank test.
USP43, ubiquitin specific peptidase 43; OS, overall survival.

Cox multivariate analysis for overall survival in 157 lung squamous cell carcinoma patients.
*Statistically significant by Cox regression test.
USP43, Ubiquitin specific peptidase 43.

Stratification survival analyses of 157 LUSC patients based on USP43 protein level.
The prognostic significance of USP43 protein expression level was evaluated in sub-grouped patients with stage T1 (A), stage T2 (B), stage T3-T4 (C), stage N0 (D), or stage N1-N2 (E). *P < 0.05 by log-rank test.
We next conducted cellular assays to validate the tumor-related role of USP43 in LUSC by knockdown strategy in NCI-H520 and NCI-H1703 cell lines (Fig. 4A-C). According to the CCK-8 assays, silencing USP43 can remarkably inhibited the proliferation capacity of both cell lines (Fig. 4D, E), which is consistent with the decreased CDK1 level (Fig. 4A-C). Similarly, colony formation experiments also revealed an impaired cell growth upon USP43-knockdown (Fig. 4F, G).
Besides proliferation, we also tested whether USP43 has any effect on LUSC migration and invasion. As revealed by Transwell migration assays (Fig. 5A, B) and Matrigel-Transwell invasion assays (Fig. 5C, D), the cells treated with USP43-shRNAs exhibited significantly decreased migration and invasion capacities, comparing with the control cells. Therefore, we finally tested several well-known biomarkers that play roles in cell invasion, and found that both vimentin and snail can be positively regulated by USP43 (Fig. 4A-C). Taken together, our cellular data demonstrated that USP43 promotes LUSC progression by enhancing proliferation and invasion.

Silencing USP43 inhibits LUSC cell proliferation.
(A, B) Western blotting analyses were conducted to test the knockdown efficiencies of shRNAs and the expression level of protein markers in NCI-H520 cells (A) and NCI-H1703 cells (B). (C) RT-qPCR assays were performed to assess the alterations of mRNA levels after knockdown in NCI-H520 and NCI-H1703 cells. (D, E) CCK-8 experiments revealed that silencing USP43 impaired the proliferation capacities of LUSC cell lines, NCI-H520 (D) and NCI-H1703 (E). (F, G) Colony formation also demonstrated the critical role of USP43 on maintaining cell proliferation of NCI-H520 cells (F) and NCI-H1703 cells (G). *P < 0.05 by Student’s t-test compared with scrambled groups.
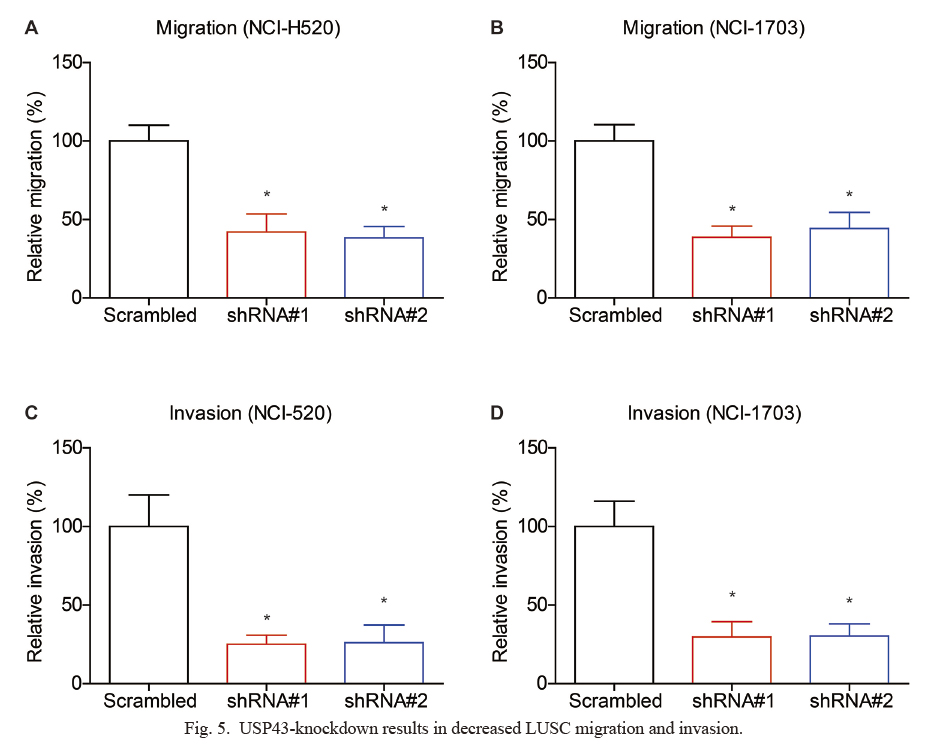
USP43-knockdown results in decreased LUSC migration and invasion.
(A, B) Transwell assays were conducted to test the migration capacities of LUSC cell lines, NCI-H520 (A) and NCI-H1703 (B), which showed that silencing USP43 attenuated the cell migration. (C, D) Matrigel-Transwell experiments revealed that USP43-shRNA can significantly inhibit LUSC invasion in NCI-H520 cells (C) and NCI-H1703 cells (D). *P < 0.05 by Student’s t-test compared with scrambled groups.
In the current study, we investigated the expression of USP43 in LUSC for the first time. Based on our data, although USP43 exhibited distinct protein levels in tissues from different patients, higher USP43 was significantly correlated with aggressive tumor characteristics such as larger tumor size and advanced TNM stages. In the enrolled retrospective cohort, higher USP43 protein expression was also correlated with poorer overall survival of LUSC. Consistently, TCGA cohort also indicated that higher USP43 mRNA transcription can help predict unfavorable LUSC prognosis. Taken together with the multivariate data, we safely came to the conclusion that USP43 can serve as a novel independent prognostic factor to help predict LUSC survival.
Since clinical data revealed the significant positive correlation between USP43 and tumor stages, we also conducted cellular assays to further validate its detailed effects in LUSC progression. Accordingly, silencing USP43 with specific shRNAs remarkably inhibited the proliferation and invasion processes of two LUSC cell lines. The effects of USP43 on LUSC growth and metastasis were consistent with its reported role in colorectal cancer and breast cancer (Lin et al. 2017; Ye et al. 2021). However, USP43 was a novel USP protein family member, only few studies reported its role in diseases. Here our data provided evidence that focusing on USP43 may be invaluable for therapeutic development.
To explore the downstream effectors of USP43, we next conducted immunoblotting assays to test the expression alteration of key regulators of proliferation and invasion. As a result, expression of CDK1, vimentin, and snail were all inhibited after silencing USP43, further confirming its oncogenic effects. Due to the equipment and technical limitations of the laboratory, we are currently unable to clarify the more detailed mechanism of USP43, such as its direct downstream substrates, which requires further research in the future (Zou et al. 2021). In addition, the combinational prognostic effect of USP43 and other biomarkers requires more clinical practice.
In conclusion, our study demonstrates the potential of USP43 as a novel prognostic biomarker for LUSC patients, and knockdown of USP43 exerts pivotal anti-cancer effects by inhibiting LUSC proliferation and invasion.
Qingchao Sun conducted clinical analyses, Haiping Zhang and Liang Zong performed cellular assays, Ainiwaer Julaiti helped collect clinical data, Xiaoliang Jing helped statistical analyses, Liwei Zhang designed this project.
The authors declare no conflict of interest.