2023 Volume 259 Issue 3 Pages 209-219
2023 Volume 259 Issue 3 Pages 209-219
The Holliday Junction-Recognition Protein (HJURP) was upregulated in several tumors, which was associated with poor outcome. This study investigated the effects of the HJURP-mediated c-Jun N-terminal kinase (JNK)/ signal transducer and activator of transcription 3 (STAT3) pathway on bladder urothelial carcinoma (BLUC). Online databases were used to analyze HJURP expression in BLUC and the correlation of HJURP to JNK1 [mitogen-activated protein kinase 8 (MAPK8)], JNK2 (MAPK9), STAT3, marker of proliferation Ki-67 (MKI67), proliferating cell nuclear antigen (PCNA), cyclin dependent kinase 2 (CDK2), CDK4 and CDK6. HJURP expression was detected in BLUC cells and human normal primary bladder epithelial cells (BdECs). BLUC cells were treated with HJURP lentivirus activation /shRNA lentivirus particles or JNK inhibitor SP600125. HJURP was upregulated in BLUC tissues and correlated with poor prognosis of patients (all P < 0.05). HJURP in tumor positively correlated with MAPK8 (R = 0.30), MAPK9 (R = 0.30), STAT3 (R = 0.15), MKI67 (R = 0.60), PCNA (R = 0.46), CDK2 (R = 0.39), CDK4 (R = 0.24) and CDK6 (R = 0.21). The JNK inhibitor SP600125 decreased p-JNK/JNK and p-STAT3/STAT3 in BLUC cells, which was reversed by HJURP overexpression (P < 0.05). The HJURP-mediated JNK/STAT3 pathway promoted BLUC cell proliferation and inhibited cell apoptosis (P < 0.05). HJURP reversed the arrested G0/G1 phase of BLUC cells by SP600125. HJURP acted as an oncogene to regulate BLUC cell proliferation, apoptosis and the cell cycle by mediating the JNK/STAT3 pathway. Therefore, HJURP targeting might be an attractive novel therapeutic target for early diagnosis and treatment in BLUC.
Bladder cancer (BCa) is the most common malignancy in the bladder mucosa of the urinary tract and one of the most prevalent cancers worldwide (Dobruch and Oszczudlowski 2021). According to estimates in 2020 (worldwide, both sexes, all ages), the incidence and the mortality of BCa were 573,278 and 212,536, respectively, which ranked 10th for malignant cancer incidence and 13th in malignancy-caused deaths worldwide. However, the incidence and the mortality of BCa were 5.9% and 2.7%, respectively, in China (http://gco.iarc.fr/today/home). BCa pathologically includes bladder urothelial carcinoma (BLUC, also known as transitional cell carcinoma), squamous cell carcinoma and adenocarcinoma, small cell carcinoma, mixed carcinoma, carcinosarcoma and metastatic carcinoma, and BLUC is the most common type, accounting for up to 95%. BLUC generally places a heavy financial burden on patients. BLUC originates from the urothelial cells on the inside of the bladder (Hyldgaard and Jensen 2021; Martinez-Rojo et al. 2021). Most BCa patients are males aged 65 years and older, and female patients are less common. The major risk factors for BCa include smoking, exposure to chemicals, radiotherapy and chemotherapy (Martinez-Rojo et al. 2021). The treatment methods of BCa are limited to surgery and immunotherapy or chemotherapy (Siracusano et al. 2020). Recent extensive analyses of molecular alterations have created the possibility of some novel therapeutic approaches for BCa (Rozanec and Secin 2020).
Holliday junction-recognition protein (HJURP), also known as FAKTS, URLC9, or HFLEG1, is located at 2q37.1, and it encodes a protein consisting of 748 amino acids. It is expressed in thymus, placenta, small intestine, liver, skeletal muscle, bone marrow and colon tissues (http://www.genecards.org/cgi-bin/carddisp.pl?gene=HJURP). HJURP is abnormally expressed in many diseases, including several cancers. For example, Tsevegjav et al. (2022) revealed that HJURP was highly expressed in most oral cancer cell lines and tissues compared to normal oral epithelial cells. Milioli et al. (2017) observed the overexpression of HJURP in basal-like breast cancer, which was linked to abnormal proliferation, cell invasion and metastasis in breast cancer and is consistent with the definition of basal-like breast carcinoma (Lehmann et al. 2011). Valente et al. (2013) reported that HJURP was overexpressed in 40 astrocytoma samples of different grades, and tumors with high HJURP expression correlated with poor survival prognosis, which indicated that HJURP overexpression was an independent prognostic factor for the risk of death of astrocytoma patients. Based on the online databases UALCAN and GEPIA, we found the upregulation of HJURP in BLUC and its positive correlation to c-Jun NH(2)-terminal kinase 1 (JNK1, also known as mitogen-activated protein kinase 8, MAPK8) (Hess et al. 2002), JNK2 (MAPK9) (Jaeschke et al. 2005), and STAT3 in BLUC. The JNK/STAT3 signaling pathway is abnormally activated in BCa, and many genes inhibit this pathway to play a potential therapeutic role in BCa (Lee et al. 2020; Mirzaei et al. 2021). Therefore, we hypothesized that HJURP mediated the JNK/STAT3 signaling pathway to exert a regulatory role in BLUC. The present study detected and compared the expression of HJURP in different BLUC cell lines and human normal primary bladder epithelial cells (BdECs). BLUC cell lines were treated with HJURP lentivirus activation particles/shRNA lentivirus particles or the JNK inhibitor SP600125, and we detected cell proliferation, apoptosis and cell cycle distribution to verify our hypothesis. The full names of abbreviations were listed in Supplementary Table S1.
The online TCGA databases UALCAN (http://ualcan.path.uab.edu/analysis.html) and GEPIA (http://gepia.cancer-pku.cn/) were used for analyses with HJURP as the keyword. HJURP expression in BLUC tumor tissues and normal tissues was analyzed and compared. Disease-free survival (DFS, also called relapse-free survival and RFS) analysis was performed using the log-rank test for hypothesis testing.
Cell cultureHuman normal primary bladder epithelial cells (BdECs) were purchased from American Type Culture Collection (PCS-420-010, ATCC, Manassas, VA, USA) and cultured in basic culture medium for bladder epithelial cells (PCS-420-032, ATCC) supplemented with a bladder epithelial growth kit (PCS-420-042, ATCC). SW780 cells (Grade I, CRL-2169, ATCC) were cultured in Leibovitz’s L-15 medium with 10% fetal bovine serum (FBS). The 5637 cells (Grade II) were cultured in RPMI-1640 medium with 10% FBS. T24 cells (HTB-4, Grade III) were cultured in McCoy’s 5A medium with 10% FBS. TCCSUP cells (Grade IV) were cultured in minimum essential medium (Eagle) in Earle’s Balanced Salt Solution (BSS) with nonessential amino acids (30-2003, ATCC) containing 1 mM sodium pyruvate (90%) and 10% FBS. The conditions for cell culture included 37°C, 95% air, and 5% CO2.
Cell treatmentT24 and TCCSUP cells were transfected with control (sc-437282)/HJURP (sc-405278-LAC) lentivirus activation particles or control (sc-437282)/HJURP (sc-94530-V) shRNA lentivirus particles (all purchased from Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) to investigate the effect of HJURP on the JNK/STAT3 signaling pathway. T24 and TCCSUP cells were treated with both HJURP lentivirus activation particles and the JNK inhibitor SP600125 (ab120065, Abcam, Cambridge, MA, USA) to examine whether HJURP overexpression reversed the inhibitory effect of SP600125 on the JNK pathway. To further analyze whether HJURP regulated the JNK/STAT3 pathway to affect the proliferation, apoptosis and cell cycle of BLUC cells, T24 and TCCSUP cells were divided into a Mock group, Control activation group, HJURP activation group, SP600125 group and HJURP activation + SP600125 group. When cell confluence reached 70-90%, transfection was performed with Lipofectamine 3000 Transfection Reagent (L3000150, Thermo Fisher Scientific Co., Ltd., Shanghai, China) in accordance with the manufacturer’s instructions. Subsequent experiments were performed 48 h after transfection.
Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)TRIzol reagent (15596026, Thermo Fisher Scientific Co., Ltd.) was used to extract total RNA from cells. A spectrophotometer was used to determine the purity and concentration of RNA. The extracted RNA was reverse transcribed into cDNA using a reverse transcription kit. A Maxima SYBR Green qPCR (K0253, Thermo Fisher Scientific Co., Ltd.) was used to analyze gene expression levels. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal reference. The 2-ΔΔCt method was used to calculate the relative expression levels of mRNA (Table 1).

Primer sequences for quantitative reverse-transcription polymerase chain reaction (qRT-PCR).
HJURP, Holliday Junction-Recognition Protein; MAPK8, Mitogen-activated protein kinase 8; MAPK9, Mitogen-activated protein kinase 9; STAT3, Signal transducer and activator of transcription 3; MKI67, Marker of proliferation Ki-67; PCNA, Proliferating cell nuclear antigen; CDK2, Cyclin dependent kinase 2; CDK4, Cyclin dependent kinase 4; CDK6, Cyclin dependent kinase 6; GAPDH, Glyceraldehyde-3-phosphate dehydrogenase.
Cells were collected for protein extraction with radioimmunoprecipitation assay (RIPA) Lysis Buffer (sc-24948, Santa Cruz Biotechnology, Inc.), and the protein concentration was determined using a BCA Protein Assay Kit (sc-202389, Santa Cruz Biotechnology, Inc.). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate proteins, which were transferred to polyvinylidene fluoride (PVDF) membranes. The PVDF membrane was blocked in Tris-buffered saline (TBS) buffer with 5% defatted milk powder for 1 h. Rabbit monoclonal antibodies were added overnight at 4°C, including an anti-HJURP antibody (ab233541, Abcam) at a 1:1,000 dilution, anti-β-actin antibody (ab8227) at a 1 µg/ml dilution, anti-JNK antibody (ab179461) at a 1:20,000 dilution, anti-JNK (phospho) antibody (ab124956) at a 1:1,000 dilution (purified), anti-STAT3 (phospho) antibody (ab76315) at a 1:20,000 dilution (purified), and anti-STAT3 antibody (ab68153) at a 1:1,000 dilution. After washing, goat anti-rabbit IgG H&L (HRP) (ab97051) at a 1:100,000 dilution was added for incubation at room temperature for 2 h. Enhanced chemiluminescence (ECL) was used for the development and visualization of protein bands. QUANTITY ONE software (Bio-Rad Laboratories, Hercules, CA, USA) was used to analyze the density of protein bands, and the experiment was repeated three times.
MTT (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay to detect cell proliferationT24 and TCCSUP cells were collected at the logarithmic growth phase and inoculated into 96-well plates at a density of 5 × 104 cells/well to culture for 24 h, 36 h, 48 h, 72 h and 96 h. Each well received 20 μL MTT (5 mg/mL) for 4 h of continued culture. The culture medium was discarded, and 150 μL dimethyl sulfoxide (DMSO) was added to each well for full dissolution and 10 min of oscillation prior to the development and measurement of absorbance at a wavelength of 490 nm.
Flow cytometry detection of cell cycle distributionCells (5 × 105 cells/well) were seeded in 6-well plates for 48 h of incubation, fixed in 80% ethanol, and centrifuged at 200 × g for 5 min before disposal of the culture medium. The cells were washed twice with 1 ml of phosphate-buffered saline (PBS), centrifuged and suspended in 100 μl of PBS. The cells were incubated in 80% ethanol for 2 h (4°C) and preserved at −20°C. The cells were stained with propidium iodide (PI), centrifuged at 200 × g for 5 min, washed twice with 1 ml PBS, and centrifuged again. The cell precipitate was incubated for 30 min (37°C) in 100 μl PBS with 1% FBS, PI (5 μg) and RNaseA (20 μg). A FACScalibur flow cytometer (Becton Dickinson, CA, USA) was used to detect the fluorescence of PI, and FlowJo software was used to analyze the data.
Annexin V-FITC/PI staining detection of cell apoptosisAfter centrifugation, cells were suspended in 200 µL PBS, followed by the addition of annexin V-FITC 10 µL and PI 5 µL for 15 min at room temperature in the dark. Binding buffer (30 µL) was added, and cell apoptosis was detected within 0.5 h. Annexin V+/PI− stained cells were counted as early apoptotic cells, and annexin V+/PI+ stained cells were late apoptotic/necrotic cells (Yu et al. 2019).
Statistical methodsGraphPad Prism 6.0 (GraphPad Software, CA, USA) was used for graphical analyses, and SPS 22.0 was used for statistical analyses. All data are presented as the means ± standard deviation (SD). One-way ANOVA was used to test the significance of mean values, and Tukey’s test was used to compare intergroup differences. P < 0.05 indicated a statistically significant difference.
To detect the expression of HJURP in BLUC, we used the UALCAN database and found that HJURP was one of the 25 overexpressed genes in BLUC (Supplementary Fig. S1). HJURP was significantly upregulated in cancerous tissues of BLUC patients (all P < 0.01, Fig. 1A, B), and it was associated with the disease-free survival of BLUC patients (log-rank P = 0.030, HR = 1.5, Fig. 1C). Patients with high HJURP expression (n = 201) had appreciably shorter DFS compared to patients with low HJURP expression (n = 201). HJURP expression in BLUC cell lines (SW780, 5637, T24 and TCCSUP) was apparently higher than BdECs (all P < 0.05, Fig. 1D, E).
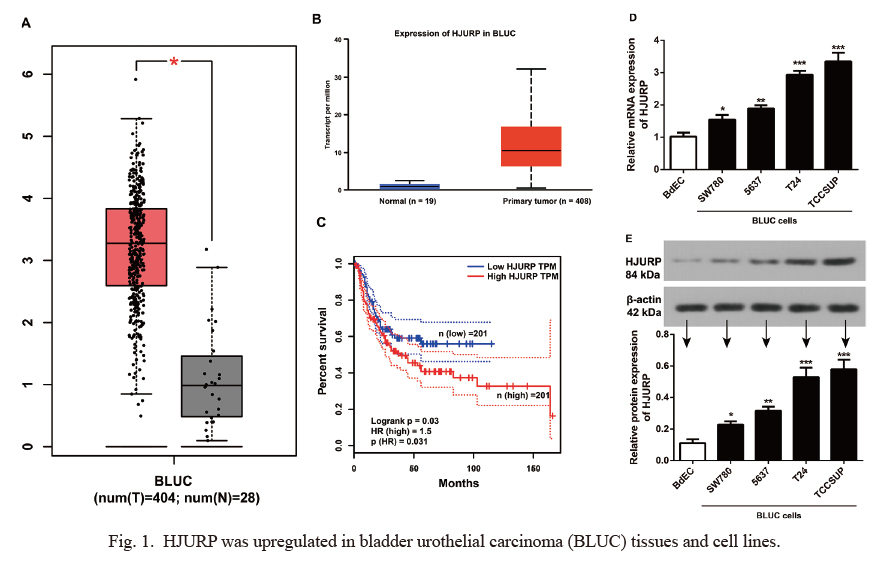
HJURP was upregulated in bladder urothelial carcinoma (BLUC) tissues and cell lines.
(A, B) The GEPIA (*P < 0.01, A) and UALCAN (P = 1.11E-16, B) databases showed upregulation of HJURP in BLUC tissues. (C) Patients with high HJURP expression had shorter disease-free survival (RFS) than patients with low HJURP expression (95% CI as dotted line) using the Cox-Mantel log-rank test. (D, E) qRT-PCR and Western blotting revealed the upregulation of HJURP in BLUC cell lines (SW780, 5637, T24 and TCCSUP) compared to BdECs (*P < 0.05, **P < 0.01, ***P < 0.005). The experiment was repeated three times.
Spearman analysis of the GEPIA database revealed that HJURP expression significantly positively correlated with JNK1 (MAPK8, R = 0.30, P = 4.5E-10, Fig. 2A), JNK2 (MAPK9, R = 0.30, P = 4.9E-10, Fig. 2B) and STAT3 (R = 0.15, P = 0.003, Fig. 2C). QRT-PCR and Western blotting were used to detect the transfection efficiency of T24 and TCCSUP cells transfected with HJURP lentivirus activation particles and HJURP shRNA lentivirus particles. HJURP gene expression was apparently upregulated and downregulated, respectively (all P < 0.05, Fig. 3A). Moreover, the gene expressions of MAPK8 (Fig. 3B), MAPK9 (Fig. 3C) and STAT3 (Fig. 3D) were positively regulated by HJURP lentivirus particles (all P < 0.05). HJURP overexpression activated the JNK/STAT3 signaling pathway with the enhanced expressions of p-JNK/total-JNK and p-STAT3/total-STAT3, and HJURP shRNA exerted the opposite effect (all P < 0.05, Fig. 3E-H). A subsequent experiment found that the JNK inhibitor SP600125 had no obvious effect on HJURP expression in BLUC cells (all P > 0.05, Fig. 3I, J), but HJURP activation particles reversed the inhibitory effect of SP600125 on p-JNK/total-JNK and p-STAT3/total-STAT3 expression in T24 and TCCSUP cells (all P < 0.05, Fig. 3K, L).
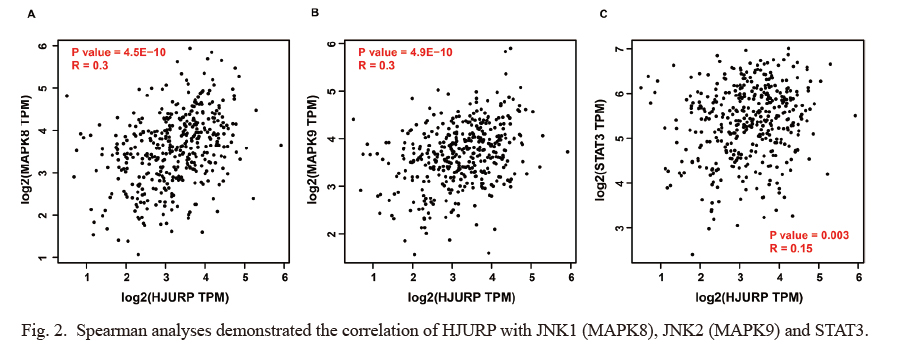
Spearman analyses demonstrated the correlation of HJURP with JNK1 (MAPK8), JNK2 (MAPK9) and STAT3.
Based on the GEPIA database, HJURP expression in BLUC tissues was in a positive correlation with the expression of MAPK8 (A), MAPK9 (B) and STAT3 (C).

HJURP regulates the JNK/STAT3 signaling pathway in BLUC.
(A-D) The expressions of HJURP (A), MAPK8 (B), MAPK9 (C) and STAT3 (D) in T24 and TCCSUP cells detected by qRT-PCR. *P < 0.05 compared to the Mock group; #P < 0.05 compared to the HJURP activation particle group; &P < 0.05 compared to the Control shRNA group. (E) Representative Western blotting for HJURP, p-JNK/total-JNK and p-STAT3/total-STAT3 protein levels. (F-H) Quantifications of HJURP/β-actin (F), p-JNK/total-JNK (G) and p-STAT3/total-STAT3 (H) in T24 and TCCSUP cells detected by Western blotting. *P < 0.05 compared to the Mock group; #P < 0.05 compared to the HJURP activation particle group; &P < 0.05 compared to the Control shRNA group. 1, Mock group; 2, Control activation particle group; 3, HJURP activation particle group; 4, Control shRNA group; 5, HJURP shRNA group. (I-L) The inhibitory effect of SP600125 on p-JNK/total-JNK and p-STAT3/total-STAT3 in T24 and TCCSUP cells was reversed by HJURP activation particles. HJURP expression in T24 and TCCSUP cells was detected by qRT-PCR (I), and quantifications of HJURP/β-actin (J), p-JNK/total-JNK (K) and p-STAT3/total-STAT3 (L) by Western blotting. *P < 0.05 compared to the Mock group; #P < 0.05 compared with the SP600125 group. The experiment was repeated three times.
Spearman analysis of the GEPIA database revealed a significant positive correlation between HJURP and the proliferation-related genes MKI67 (R = 0.60) and PCNA (R = 0.46) (Fig. 4A). The MTT assay found that HJURP upregulation led to significantly increased proliferation of BLUC cells, and SP600125 apparently inhibited the proliferation of BLUC cells. Compared to the SP600125 group, the HJURP activation + SP600125 group showed increased cell proliferation and a significant upregulation of MKI67 and PCNA expression (all P < 0.05, Fig. 4B, C).

HJURP mediates the JNK/STAT3 signaling pathway to affect BLUC cell proliferation.
(A) Spearman analysis of the GEPIA database revealed a significant positive correlation of HJURP with MKI67 and PCNA. (B) Proliferation of BLUC cells evaluated using the MTT assay. (C) Gene expression of MKI67 and PCNA in T24 and TCCSUP cells detected by qRT-PCR. *P < 0.05 compared to the Mock group; #P < 0.05 compared to the HJURP activation particles group; &P < 0.05 compared to the SP600125 group. The experiment was repeated three times.
As shown in Fig. 5A, B, SP600125 arrested T24 and TCCSUP cells at the G0/G1 phase and reduced the proportion of S phase cells, which was reversed by HJURP overexpression particles. Analysis of the GEPIA database found that HJURP significantly positively correlated with the G1 phase cycle-related genes CDK2 (R = 0.39, P = 4.4E-16), CDK4 (R = 0.24, P = 1.5E-06) and CDK6 (R = 0.21, P = 1.5E-05) (Fig. 5C). HJURP overexpression promoted the expression of CDK2, CDK4 and CDK6 in T24 and TCCSUP cells (all P < 0.05), and SP600125 reduced the expression of CDK2, CDK4 and CDK6 in BLUC cells (all P < 0.05). Compared to the SP600125 group, T24 and TCCSUP cells in the HJURP activation + SP600125 group showed markedly increased expression of CDK2 and CDK4 (all P < 0.05, Fig. 5D).

HJURP mediates the JNK/STAT3 signaling pathway to affect BLUC cell cycle distribution.
(A) Cytometrical analysis quantified the cell cycle distributions of T24 and TCCSUP cells. Hatched blue peaks denote G0/G1 (left) and G2/M (right) phases. Hatched fuchsia areas denote S phase (middle). (B) SP600125 arrested T24 and TCCSUP cells at the G0/G1 phase and reduced the proportion of S phase cells, which was reversed by HJURP overexpression particles. (C) Spearman analysis of the GEPIA database revealed a significant positive correlation of HJURP with the G1 phase cycle-related genes CDK2, CDK4 and CDK6. (D) Gene expression of CDK2, CDK4 and CDK6 in T24 and TCCSUP cells detected by qRT-PCR. *P < 0.05 compared to the Mock group; #P < 0.05 compared to the HJURP activation particles group; &P < 0.05 compared to the SP600125 group. The experiment was repeated three times.
Annexin V-FITC/PI staining was performed to detect cell apoptosis (Fig. 6). Compared to the Mock group, HJURP significantly reduced the apoptosis rate of BLUC cells (T24 and TCCSUP), and SP600125 appreciably increased the cell apoptosis rate (all P < 0.05). HJURP overexpression effectively reversed the proapoptotic effect of SP600125 on BLUC cells, which was demonstrated by the dramatic decrease in the number of apoptotic cells in the HJURP activation + SP600125 group compared to the SP600125 group (all P < 0.05).

Annexin V-FITC/PI staining detects the effect of the HJURP-mediated JNK/STAT3 signaling pathway on the apoptosis of BLUC cells (T24 and TCCSUP).
(A) Representative plots of showing Annexin V-FITC/PI staining. (B) The apoptotic rate was calculated by adding the percentages of two quadrants, including lower right quadrant (early-stage apoptotic cells, Annexin V-FITC+/PI−) and upper right quadrant (late-stage apoptotic cells, Annexin V-FITC+/PI+). *P < 0.05 compared to the Mock group; #P < 0.05 compared to the HJURP activation particles group; &P < 0.05 compared to the SP600125 group. The experiment was repeated three times.
HJURP was upregulated in BLUC tissues, which was correlated with the poor prognosis of patients and the expression MAPK8, MAPK9 and STAT3 by online database. This was the first study which revealed that decreased p-JNK/JNK and p-STAT3/STAT3 by a JNK inhibitor SP600125 in BLUC cells was reversed after transfection with HJURP activation lentivirus particles. The HJURP-mediated JNK/STAT3 pathway could affect BLUC cell proliferation, cell apoptosis, and cell cycle.
HJURP, a centromeric histone chaperone involved in de novo histone H3 variant CenH3 (CENP-A) recruitment (Niikura et al. 2015), was one of the 25 overexpressed genes in BLUC based on biological information analyses, and HJURP expression was significantly higher in BLUC cell lines (SW780, 5637, T24 and TCCSUP) than BdECs. Similar expression of HJURP was also observed in many other cancers (Dou et al. 2022; Jia et al. 2022; Tsevegjav et al. 2022). Notably, the mRNA and protein levels of HJURP were significantly upregulated in BCa tissues in a transcriptome and in vivo study (Cao et al. 2017), which is consistent with the findings in the current study. The overexpression of HJURP in hepatocellular carcinoma (HCC) tissues primarily resulted from the hypomethylation of the HJURP promoter region (Li et al. 2021). However, whether the upregulation of HJURP in BLUC was also affected by methylation was not known, and further investigations are needed. As a DNA repair protein, HJURP is associated with the proliferative competence, drug resistance and poor prognosis of cancer cells (Sousa et al. 2019). High HJURP expression predicts a higher risk of metastatic relapse and poor outcome in breast cancer patients (Montes de Oca et al. 2015). Zhang et al. (2021) also revealed that the prognostic factor HJURP influenced the prognosis of non-small-cell lung cancer (NSCLC) patients. BLUC patients with high HJURP expression had apparently shorter disease-free survival than patients with low HJURP expression, which suggests that HJURP acts as an oncogene in bladder cancer and affects the prognostic outcome of patients.
Based on the Spearman analysis of the GEPIA database, HJURP significantly positively correlated with JNK1 (MAPK8), JNK2 (MAPK9) and STAT3. A previous study demonstrated that JNK activation was essential for tobacco smoke-induced lung cancer, chemical carcinogen-induced gastric cancer, hepatocellular carcinoma (Liu et al. 2012), and BLUC (Shen et al. 2016) in animal models. STAT3 mediates many cell processes, including the metabolism, survival, host defense, growth and differentiation, and it is unremittingly activated in cancer cells compared to normal cells (Saleem et al. 2020). STAT3 activation upregulates the expression of genes related to angiogenesis, metastasis and proliferation (Cui et al. 2020), and inhibition of STAT3 activation may be a novel approach to inhibit tumor growth (Saleem et al. 2020). The carcinogenic effect of the JNK/STAT3 signaling pathway was found in many cancers, such as epithelial ovarian cancer, lung cancer, and oral squamous cell carcinoma (OSCC) (Gkouveris et al. 2017; Cui et al. 2020; Wang et al. 2020). A previous study reported that ectopic HJURP expression promoted the proliferation of prostate cancer cells in vitro and in vivo via the JNK signaling pathway (Lai et al. 2021). Based on the online analysis, we found that HJURP expression in BLUC tissues was in a positive correlation with the expression of MAPK8, MAPK9 and STAT3. In addition, HJURP knockdown decreased gene expressions of MAPK8, MAPK9 and STAT3 in BLUC cells with decreased ratio of p-JNK/JNK and p-STAT3/STAT3, while the HJURP overexpression caused opposite effect. All mentioned above indicated that HJURP may target the transcription of JNK/STAT3, thus inducing protein degradation. We treated BLUC cells with the JNK pathway inhibitor SP600125 (Bennett et al. 2001; Pereira et al. 2012) and found that SP600125 had no obvious effect on HJURP expression. However, HJURP overexpression counteracted the inhibitory effect of SP600125 on the JNK pathway, which indicates that HJURP may play a regulatory role in BLUC by mediating the JNK signaling pathway.
The histone chaperone HJURP mediates the deposition of CENP-A by colocalizing with CENP-A at the centromere in early G1 phase (Andronov et al. 2019). Online database analyses found a significant positive correlation between HJURP and the G1 phase cycle-related genes CDK2, CDK4 and CDK6. A previous study showed that low HJURP expression arrested cancer cells at the G1 phase (Filipescu et al. 2017), including serous ovarian cancer cells (Dou et al. 2022). U343MG cells with conspicuous senescence were significantly arrested at the G1 phase with a decreased proportion of S phase cells on the fifth day after HJURP suppression (Serafim et al. 2020). SP600125 inhibited cell growth by arresting cells at the G1 or G2/M phase in multiple myeloma (MM) cell lines (Hideshima et al. 2003). The inhibitory effect of SP600125 on human mast cell proliferation was also related to cell cycle arrest at the G1 phase (Wang et al. 2006). SP600125 arrested BLUC cells at the G0/G1 phase and reduced the expression of CDK2, CDK4 and CDK6 in the present study, which was reversed by HJURP overexpression. Inhibition of HJURP expression reduced the proliferative competence of four colorectal cancer (CRC) cell lines in vitro (Kang et al. 2020). HJURP knockdown downregulated clonogenic capacity and severely damaged the survival of five distinct glioblastoma (GBM) cell lines (Serafim et al. 2020). We also found that HJURP overexpression reversed the inhibitory effect of SP600125 on the proliferation of BLUC cells, which was accompanied by increased cell apoptosis and cell cycle alterations. We performed Spearman analysis using the GEPIA database and found a significant positive correlation between HJURP and cell proliferation markers (Helpap et al. 2003), including MKI67 and PCNA. A similar trend was reported in breast cancer, where the HJURP mRNA level significantly correlated with the proliferation index MKI67 (Hu et al. 2010). HJURP overexpression also upregulated the expression of MKI67 and PCNA in BLUC cells in our study, and SP600125 reduced the expression of MKI67 and PCNA. These results suggest that HJURP mediates the JNK/STAT3 signaling pathway to affect the proliferation, apoptosis and cell cycle distribution of BLUC cells.
There are some potential limitations of our study. First, this was an in vitro cell study, therefore results shown here do not necessarily replicate the effects of human tumor microenvironment. Secondly, we did not evaluate the underlying molecular mechanisms of HJURP-mediated JNK/STAT3 signaling pathway in BLUC in vivo. Thirdly we tested only 2 cell lines and it is not known how other BLUC cell lines would respond to SP600125.
In summary, HJURP was upregulated in BLUC tissues and cell lines and acted as an oncogene to promote proliferation, inhibit apoptosis, and regulate the cell cycle distribution of BLUC cells via regulation of the JNK/STAT3 signaling pathway. Findings of our study theorize that HJURP is a novel protein which has an important role in progression of BLUC possibly associated with JNK/STAT3 signaling pathway, and it might be an attractive novel therapeutic target for its early diagnosis and treatment.
This work was supported by the Zhejiang University Cooperation Project of Lishui (No. 2018ZDHZ12), Zhejiang Public Welfare Technology Application Research Project (No.LGF20H050003), Zhejiang Medical and Health Science and Technology Plan Project (No. 2020382934), Traditional Chinese medicine of Zhejiang province science and technology plan project (No. 2020ZB309), Lishui Public Welfare Technology Application Research Project (No. 2019SJZC46), and Lishui Public Welfare Technology Application Research Project (No. 2020GYX22).
The authors declare no conflict of interest.