2014 Volume 37 Issue 6 Pages 1068-1074
2014 Volume 37 Issue 6 Pages 1068-1074
Tribbles related protein 3 (TRB3) pseudokinase plays a crucial role in cell proliferation, migration and morphogenesis during development. In our recent study, an introduction of human TRB3 gene into mouse mammary tumor cells caused an increase of proliferation of tumor cells and their nuclear size. In the current study, to examine whether this gene causes de novo morphological changes in a specific organ site we have developed a novel variation of the transgenic mouse model that conditionally expresses human TRB3 (hTRB3) gene using Cre-recombinase (Cre)/loxP recombination system. By injecting hTRB3 transgene construct into pronuclei of mouse embryo, we eventually obtained four hTRB3 mice. The gene expression was controlled by infection of adenovirus-expressing Cre via the tail vein of hTRB3 mouse. In Cre-mediated hTRB3 mouse, expression of the hTRB3 protein was detected in the cytoplasm of hepatocytes in the liver. Expression of this protein was also seen in lymphocytes in the spleen, glomerular endothelial cells, and epithelial cells of collecting duct of the kidney. In hepatocytes of the hTRB3 mouse, nuclear size was significantly greater than that of the wild type mouse, indicating that hTRB3 can play a role at least in part in hepatic morphogenesis. The present animal model may provide a system for evaluation of de novo morphological changes induced by a specific transgene in a specific organ site.
Tribbles related protein 3 (TRB3) is one of the mammalian homologues of Drosophila Tribbles. This molecule contains a serine/threonine kinase catalytic domain but lacks an ATP binding site or one of the conserved catalytic motifs essential for kinase activity.1–3) TRB3 has also been shown to be involved in multiple cellular processes such as glucose/lipid metabolism, muscle/adipocyte differentiation, and stress response by interacting with various functional proteins.1,3–10) Three mammalian Tribbles homologues, TRB1, TRB2, and TRB3 are crucial modulators of tumorigenesis.3,11–13) TRB1 and TRB2 induce acute myelogenous leukemia by inhibiting CCAAT/enhancer binding protein α (C/EBPα) function.12) TRB3 is highly expressed in a wide range of human carcinoma cell lines and in several types of human carcinoma.3,13) We have recently demonstrated that TRB3 promotes proliferation and induces polyploidy of mouse mammary tumor cells.14) This finding was likely to indicate TRB3’s response to morphological function. In the current study, by utilizing the recombinant adenovirus expressing Cre-recombinase (Cre) system we examined whether human TRB3 gene causes de novo morphological changes in a specific organ site.
We ligated flag-tagged full length human TRB3 (hTRB3) cDNA7) into KpnI and SwaI restriction sites of pCALNL5 vector (DNA Bank, RIKEN Bio Resource Center, Ibaraki, Japan),15–17) and this construct was termed pCALNL-flag-hTRB3. The pCALNL-flag-hTRB3 was digested at SspI/HindIII restriction sites.
African green monkey kidney fibroblast cell line COS7 was obtained from Cell Resource Center for Biomedical Research Institute of Development, Aging and Cancer, Tohoku University, Sendai, Japan and cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Wako Pure Chemical Industries, Ltd., Osaka, Japan) containing 10% fetal bovine serum (FBS) (Life Technologies, Inc., Rockville, MD, U.S.A.). Expression vectors pCALNL-flag-hTRB3 (generated in this study), pcDNA3.1-flag-hTRB3 (used as positive control),7) pxCANCre (DNA Bank, RIKEN Bio Resource Center) and empty vector pCALNL5 (used as negative control) were transfected into COS7 cells using FuGENE6 reagent (Roche Diagnostics Corp., Indianapolis, IN, U.S.A.), and these cells were cultured in in a 6-cm culture dish. Using the transfected cells, levels of protein expression were confirmed by Western blot assays.
After confirming the Cre/loxP system functions by examining the expression status of hTRB3 that can be measured with Flag expression, the purified cassette (Fig. 1) was injected into the pronuclei of C57BL/6NCrSlc mouse (CLEA Japan, Tokyo, Japan). A total of 184 injected eggs were transplanted into pseudo-pregnant C57BL/6NCrSlc mouse. Of 8 potential transgenic mice screened, four mice were shown to carry the transgene by polymerase chain reaction (PCR) as described below. These four transgenic founder mice were then mated with wild type C57BL/6NCrSlc mice (wild mice) and offspring were screened for the presence of the transgene by PCR assay of genomic DNA isolated from tail biopsies at the age of 3 weeks. The following primers were used: geno-hTRB3F (5′-TGT CTG GAT CAA ATC CGA AC-3′) and geno-hTRB3R (5′-ATC CGT CGA CCC TTG TCA TC-3′). In the present study, we used these transgenic mice termed NCrSlcBL6-TgN(hTRB3) mice (hTRB3 mice). The mouse was maintained in plastic cages in an airconditioned room with a 12 h light/12 h dark cycle.

The pCALNL-flag-hTRB3 construct is comprised of a hybrid CMV enhancer/chicken β-actin (CAG) promoter, a cassette for the neomycin resistance gene flanked by loxP sites, and a sequence containing a human TRB3 with a Flag-tag. Infection with the Cre expressing adenovirus results in recombination of hTRB3 construct, eventually generating a functional flag-hTRB3 gene expression unit. GpA, rabbit β-globin poly A site; pA, SV40 early poly A site.
hTRB3 mouse embryonic fibroblast cell line (hTRB3 MEF) that carries human TRB3 gene was generated from hTRB3 mouse. MEF cell line was also generated from wild mouse. The hTRB3 female mouse was euthanized at 14 d postcoitum and embryos were removed from placenta. After excising head and viscera, the embryo was cut into small pieces until it became possible to pipette. One milliliter of trypsin/EDTA (Life Technologies) was added to one embryo and incubated at 37°C for 15 min. After adding DMEM containing 10% FBS, centrifugation was done and supernatant containing MEF cells was used for cell culture. MEF cell lines were cultured in DMEM containing 10% FBS.
To see whether the Cre/loxP system properly functions, we examined the expression status of mRNA and protein of hTRB3 in MEF cells. Adenovirus carrying the Cre gene (AxCANCre) was prepared as described previously.15,16) Adenovirus vectors were amplified in HEK-293 cell line that was purchased from ATCC (Manassas, VA, U.S.A.), cultured in DMEM containing 10% FBS, and purified using Vivapure Adenopack (Vivascience, Hannover, Germany). The titer of the adenovirus was determined by using the Rapid titer kit (Clontech, Mountain View, CA, U.S.A.). The virus stock was concentrated to 1.0×1010 pfu per mL. MEF cells with or without hTRB3 transgene were infected with Cre expressing adenovirus (0, 100, 200 multiplicity of infection (MOI)) for 2 h. After the infection, cells were cultured in DMEM containing 10% FBS for 48 h, genomic DNA, total RNA, and protein were extracted and used for PCR, reverse transcription (RT)-PCR, and Western blot assays.
After confirming that the Cre/loxP system functions in vitro, the system was introduced into mice and initiated by infecting animals with Cre expressing adenovirus. Cre expressing adenovirus (1×108 pfu/mouse) was injected into the tail vein of 4-week-old mouse. To induce Flag-tagged-human TRB3 gene, adenovirus in which the expression of Cre was under the control of the CAG promoter was used. Ten days after Cre expressing adenovirus was injected, all mice were euthanized and complete autopsy was done. Liver, spleen, and kidneys were carefully removed and processed for macroscopic examination and histopathological/immunohistochemical analyses. All experimental procedures conformed to the Regulations for Animal Experimentation at Nagoya City University were reviewed by the Institutional Laboratory Animal Care and Use Committee of Nagoya City University and finally approved by the President of University.
Immunohistochemical AnalysisThese assays were performed using an established method as described elsewhere.18) Three-micrometer-thick paraffin sections were prepared from liver, spleen and kidney. These sections were treated in 3% H2O2 for 10 min to block the endogenous peroxidase activity. For antigen retrieval, the sections were brought to boiling in 0.1 M citrate buffer, pH 6.0. Sections were then incubated with a primary antibody (1 : 500 dilution) of the FLAG (Sigma, St. Louis, MO, U.S.A.) at room temperature for 1 h. After incubation with secondary antibody, sections were then stained using an ABC kit (Vector Laboratories Inc., Burlingame, CA, U.S.A.) according to the manufacturer’s instructions. The number of Cre-treated wild mouse was one, and that of Cre-treated hTRB3 mice was eight. The number of untreated wild mouse was one and that of untreated hTRB3 mouse was also one. Each liver slide has two to four tissue slices. In the liver tissue slices of hTRB3 and wild mice, the longest diameter of nucleus of the liver cell was determined by image analysis (Olympus DP70 system, Olympus Corp., Tokyo, Japan). Four high power fields per liver tissue were examined and more than 100 nuclei were counted in each liver tissue.
RT-PCR and PCR AssaysThese assays were done with established procedures.18) Total RNA was extracted from each cell line grown in 9-cm culture dishes using ISOGEN (Nippon Gene, Toyama, Japan). The reaction mixture contains 4 µg of total RNA, 1 µL of 10 mM deoxyribonucleotide triphosphate (dNTP) (Life Technologies), 1 µL of Random primers (Life Technologies) and 7 µL of distilled water. The reaction mixture was then incubated at 65°C (5 min) for denaturation, chilled on ice for 1 min and added 4 µL of 5×RT buffer (Life Technologies), 1 µL of 0.1 M dithiothreitol (DTT), 1 µL of the RNase out (Life Technologies) and 1 µL of Superscript III Reverse Transcriptase (Life Technologies). After the addition of these reagents, the reaction mixture was incubated at 50°C (1 h) for random primer annealing and 70°C (15 min) for cDNA preparation. One microliter of the reaction mixture was then used for PCR. The genomic DNA was isolated from MEF cells that were derived from hTRB3 and wild mice. The primer sequences of PCR analysis used in this study were as follows: human TRB3-specific primer set, hTRB3F (5′-CAA GTC GCT CTG AAG GTT CC-3′) and hTRB3R (5′-CCA TCC TAC TCT GGC AAA GC-3′), Cre/loxP recombination primer set, CAGpF (5′-CGT GCT GGT TGT TGT GCT GTC T-3′), and geno-hTRB3R (5′-ATC CGT CGA CCC TTG TCA TC-3′). β-Actin-specific DNA fragments from the same RNA samples were amplified and served as internal controls. Primers actin F (5′-CCG TAA AGA CCT CTA TGC CAA CA-3′) and actin R (5′-CGG ACT CAT CGT ACT CCT GCT T-3′) were used for amplification of β-actin. Interleukin-2 (IL-2)-specific DNA fragments from genomic DNA samples were also amplified and served as internal controls. Primers IL-2 F (5′-CTA GGC CAC AGA ATT GAA AGA TCT-3′) and IL-2 R (5′-GTA GGT GGA AAT TCT AGC ATC ATC C-3′) were used for amplification of IL-2. PCR was conducted for 26–30 cycles in an iCycler (Bio-Rad Laboratories, Inc., Hercules, CA, U.S.A.). Each amplification cycle consisted of at 94°C (0.5 min) for denaturation, 60°C (0.5 min) for primer annealing, and 72°C (1 min) for extension. After PCR amplification, the DNA fragments were stained with ethidium bromide and analyzed by 2% agarose gel electrophoresis. The results were confirmed by repeating experiments.
Western Blot AssaysThese assays were done with established procedures.19) The cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 0.1% sodium dodecyl sulfate (SDS), 0.5% deoxycholate, and 1% Triton X-100). The lysates were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) (12.5%), transferred onto a polyvinylidene difluoride (PVDF) membrane (Immobilon P, Millipore Corp., Bedford, MA, U.S.A.) and probed with the antibodies. The primary antibodies used in the present study were anti-β-actin monoclonal antibody (AC-15, Sigma, St. Louis, MO, U.S.A.) and anti-FLAG monoclonal antibody (M2, Sigma, St. Louis, MO, U.S.A.). The antiserum against human TRB3 was prepared as described previously.7) Proteins of interest were visualized using immunoStar Zeta (Wako) and light emission was quantified with a Light-capture (ATTO Corp., Tokyo, Japan). Each assay was repeated more than three times to confirm the results.
Statistical AnalysisDifferences in the mean diameter of the nucleus of the liver cells between hTRB3 and wild type mice were analyzed by Student’s or Welch’s t-test. The value of p<0.05 was considered to be significant.
We have developed a novel variation of the transgenic mouse model by using Cre/loxP system to express TRB3 protein in the liver, spleen, and kidney tissues. Mean diameter of nucleus was greater in hepatic cells transfected with hTRB3 than non-transfected hepatocytes. This morphological difference depending on transfection status is in accordance with our recent experiments demonstrating that hTRB3 transfected mouse mammary tumor cells exhibited enlarged nuclear size and that TRB3 gene promotes cell proliferation and chromosomal instability by causing polyproidization during development of mouse mammary tumor.14) Taken together, hTRB3 gene may influence its response to morphological function and the present animal model system would be useful to see if this gene causes de novo morphological changes leading to tumorigenesis in a specific organ site.
Conditional Expression of Human TRB3 (hTRB3) Transgene Was Controlled by Cre-RecombinaseThe system was originated by investigators.15,20) This system is a powerful on/off switching tool when timed/tissue specific expression of gene is critical in cultured cells or transgenic animals. This system can also be used in experiments of developmental processes involving proteins that are suspected of exerting different functions in embryogenesis and in adult animals and that are lethal if the gene is expressed in the embryo. In the present study, little was known about the lethality when hTRB3 gene was introduced into mouse embryo. Thus, we selected this Cre/loxP system to avoid the issue of embryonic lethal.
In Fig. 2 panel A, lysate sample in lane 1 was derived from COS7 cells that were transfected with both hTRB3 transgene construct (pCALNL-flag-hTRB3) and Cre expression vector (pxCANCre). Lysate samples of lanes 4 and 5 were negative and positive controls, respectively. When COS7 cell were transfected with both hTRB3 transgene construct and Cre expression vector, the FLAG expression was present (lane 1) as seen in the positive control (lane 5). Negative and positive controls (lanes 4 and 5) consisted of COS7 cells transfected with empty vector (pCALNL5) and hTRB3 expression vector (pcDNA3.1-flag-hTRB3), respectively. These findings indicate that expression of hTRB3 transgene was controlled by the Cre/loxP system. This hTRB3 construct was used for injection as described below.

(A) Conditional expression of hTRB3. Lysate sample in lane 1 was derived from COS7 cells that were transfected with both hTRB3 construct (pCALNL-flag-hTRB3) and Cre expression vector (pxCANCre). Lysate samples of lanes 2 and 3 consisted of COS7 cells transfected with hTRB3 construct and Cre expression vector, respectively. Lysate samples of lanes 4 and 5 were negative and positive controls, respectively. Negative and positive controls (lanes 4 and 5) consisted of COS7 cells transfected with empty vector (pCALNL5) and hTRB3 expression vector (pcDNA3.1-flag-hTRB3), respectively. The FLAG expression was present (lane 1) as seen in the positive control (lane 5). (B, C) Detection of hTRB3 DNA fragments by PCR assay. We used hTRB3 transgenic mice (hTRB3 mice) as described in Materials and Methods. Genomic DNA was extracted from mice tails and used for amplification of hTRB3 and IL-2 DNA fragments. (B) hTRB3 DNA fragment was seen in 4 mice of 8 potential transgenic mice. The number indicates the individual mouse number. (C) hTRB3 DNA fragment was seen in 7 of 8 offspring generated by mating hTRB3 mice with wild mice.
Wagner et al. previously reported that the ratio of the number of offspring/embryos to foster mother was 23%.21) The ratio of the number of mouse that expresses transgene/the number of offspring was 10–40%.22)
In the present Cre/loxP system, the ratio of the number of mouse expressing transgene/embryos to foster mother was 4/184=2%. After infection of Cre, this enzyme excises the stuffer DNA region loxP–neo-poly A–loxP (Fig. 1). This step initiates gene expression of hTRB3. By introducing hTRB3 transgene construct (Fig. 1) into pronuclei of mouse embryo, four hTRB3 mice were eventually obtained (Fig. 2 panel B). These four hTRB3 mice were mated with wild mice and offspring were produced. hTRB3 genotyping was done using genomic DNA derived from the offspring, and most of them carried hTRB3 (Fig. 2 panel C). In three of four mice, hTRB3 gene was transmitted to next generation.
To confirm whether Cre-mediated recombination of the transgene occurs in MEF cells, we examined the existence of DNA fragment consisting of hTRB3 construct by PCR assay. hTRB3 MEF cells and MEF cells were generated from 14-d-old embryos of hTRB3 and wild mice, respectively. Cells were then infected with Cre expressing adenovirus as described in Materials and Methods. A 168-bp DNA fragment was seen when the recombination occurred (Fig. 3 panel A). However, a longer 1374-bp DNA fragment was seen unless hTRB3 MEF cells were treated with Cre expressing adenovirus, indicating no recombination of the transgene. We also examined the mRNA/protein expression status of hTRB3 by RT-PCR/Western blot assays. There was a marked mRNA/protein expression of hTRB3 in hTRB3 MEF cells treated with Cre (Fig. 3, panels B left and C left). There was no expression seen in MEF cells that were treated with Cre (Fig. 3, panels B right and C right).

hTRB3 MEF and MEF cells were generated from hTRB3 and wild mice, respectivly. These cells were treated with Cre expressing adenovirus (0, 100, 200 MOI). MOI indicates multiplicity of infection. (A) Confirmation of recombination of hTRB3 construct in hTRB3 MEF cells by PCR assay. A 168-bp DNA fragment derived from recombinant hTRB3 construct was seen in cells treated with Cre expressing adenovirus. hTRB3 DNA fragments were not PCR-amplified in MEF cells (data not shown). (B) Confirmation of hTRB3 mRNA expression in hTRB3 MEF/MEF cells by RT-PCR assay. Increasing mRNA expression was seen in hTRB3 MEF cells (left panel). No mRNA expression was seen in MEF cells (right panel). (C) Confirmation of hTRB3 protein expression by Western blot assay. hTRB3 protein expression was observed in hTRB3 MEF cells but not in MEF cells. β-Actin was used as an internal control.
To examine whether hTRB3 gene causes de novo morphological changes leading to tumorigenesis in a specific organ site, we performed immunohistochemical analysis and hematoxylin/eosin staining for histopathological examination in liver, spleen, and kidneys of the hTRB3 mouse. There were no apparent abnormalities seen in morphology and behavior in either hTRB3 or wild mice. As shown in Fig. 4, hTRB3 and wild mice, both of which were not infected with Cre expressing adenovirus, exhibited no significant histopathological differences in the liver (Figs. 4A, D), spleen (Figs. 4B, E), and kidney (Figs. 4C, F) tissues. There were also no apparent histopathological differences in the spleen and kidney tissues between hTRB3 and wild mice, both of which were infected with Cre expressing adenovirus (Figs. 5B, E, and Figs. 5C, F). In the hTRB3 mouse that infected with Cre expressing adenovirus, approximately 20% of hepatocyets exhibited positive cytoplasmic staining of FLAG, indicating hTRB3 protein was expressed in these hepatocytes (Fig. 6A, arrow), whereas the remaining 80% stained faintly or uncertainly. Positive FLAG staining was also seen in sinusoid in liver tissue (Fig. 6A arrowhead). In the spleen of the hTRB3 mice, approximately 10% of lymphocytes were positively stained with FLAG (Fig. 6B). In the kidney of the hTRB3 mice, approximately 20% of epithelial cells of the tubules and collecting duct were positively stained with FLAG (Fig. 6C). Also, endothelial cells of capillaries of glomeruli were positive (Fig. 6D). Perivascular inflammation in liver tissue was also seen in the hTRB3 mouse (Fig. 5A). In liver cells of the hTRB3 mouse, the mean longest diameter of nucleus was significantly greater than that of the wild mice (9.4±0.19 versus 6.3±0.11 µm, p<0.001) (Fig. 5A versus 5D). These results indicate that the Cre/loxP recombination system functions at least in part in liver, spleen, and kidney of the C57BL/6NCrSlc mouse, and that hTRB3 gene causes morphological changes in nuclear size of hepatocytes. An increase in nuclear size occurred in both FLAG-positive and negative hepatocytes (Figs. 5A, D). This is presumably because endogenous TRB3 in mouse hepatocytes may function as exogenous hTRB3 simultaneously does. Indeed, in a recent experiment we found that both endogenous and exogenous TRB3 express in a cell line that stably expresses exogenous TRB3 gene.14) Further, Bowers et al. postulate that there is a feedback response of TRB3 protein accumulation activating a higher expression of TRB3 transcript.3)

Note that no apparent histological differences were observed between hTRB3 (A–C) and wild (D–F) mice, both of which were not introduced hTRB3 transgene. Magnification was ×400 in all six images.

hTRB3 and wild mice were infected with Cre expressing adenovirus and HE staining was conducted with liver, spleen and kidney tissues derived from hTRB3 mouse (A–C) or wild mouse (D–F). No apparent abnormality was seen in liver tissue derived from wild mouse (B). Note that nuclear size of hepatocyte in the liver tissue derived from hTRB3 mouse increases compared to that of wild mouse (A). Slight perivascular inflammation is also seen (A). Note that no apparent histological differences in spleen or kidney tissues were observed between hTRB3 (B, C) and wild (E, F) mice. Magnification was ×400 in all six images.
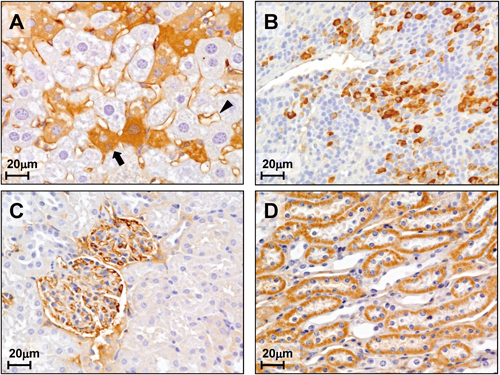
hTRB3 and wild mice were infected with Cre expressing adenovirus and immunohistochemical staining of liver (A), spleen (B) and kidney (C, D) tissues derived from hTRB3 mouse were done. (A) Immunohistochemical staining of liver tissue derived from hTRB3 mouse. Note that FLAG-mediated hTRB3 protein expression was seen in the cytoplasmic region of hepatocytes (arrow) and sinusoid of the liver tissue (arrowhead). FLAG-mediated hTRB3 protein expression was faintly or uncertainly stained in the remaining hepatocytes. (B) FLAG-mediated hTRB3 protein expression was seen in lymphocytes of spleen tissue. (C) In the kidney tissue of the hTRB3 mouse, hTRB3 was positively stained in epithelial cells of the tubules and collecting duct. (D) hTRB3 was also positively stained in endothelial cells of the capillary of glomeruli of the kidney. Magnification was ×400 in all six images.
We first estimated that Cre-mediated recombination of the transgene had occurred more than 50% in frequency. Anton et al. also estimated approximately 50% of Cre-dependent recombination in vitro.20) Kudoh et al. developed a mouse model for Duchenne muscular dystrophy using a Cre/loxP recombination system, and the ratio of the number of 100% chimeras/the number of embryo transplanted ranged from zero to 5%.23) In our experiment, actual efficiency of the present system resulted in 20%. Differences in experimental systems (i.e., in vitro/in vivo, animal strain used in the experiment, sequence of the restrictive target sites, and MOI) may explain discrepancies reported on efficiency of recombination. It may be possible for us to improve on this efficiency by optimizing MOI values or the timing of adenovirus infection.
Commonly used adenoviruses derived from human serotype 2 (Ad2) and 5 (Ad5) infect a broad range of organ tissues through interaction with the 46-kDa coxackie and adenovirus receptor (CAR).24) The major issue for successful site specific transgene delivery is tissue selectivity. Systemically injected adenovirus into mice primarily localize to hepatocytes with consequent hepatotoxicity.24–26) In the present study, we used adenovirus vector Ad5 and FLAG-mediated expression of hTRB3 transgene was seen in three organs such as liver, spleen, and kidneys, indicating low tissue selectivity.
In the hTRB3 mice, we found that nuclear diameter of FLAG-stained hepatocytes was significantly greater than that of wild mice. Some investigators report an increase in nuclear size as the disorder progresses from normal to carcinoma.27,28) Furthermore, nuclear size increases monotonically with nuclear DNA content.29) The findings in the present study together with our recent experiment14) indicate that TRB3 affects nuclear size of hepatocytes that were infected with Ad5. There are several documents demonstrating morphological changes and hepatocarcinogenesis. Large cell change of hepatocytes was regarded as a precancerous lesion.30) Nuclear enlargement of hepatocytes was induced by the treatment of animals with carcinogen.31–33) Of these reports, Clawson et al. found that hepatocarcinogen induced nuclear enlargement is associated with substantial diploid to tetraploid shifts in hapatocytes.31) Regarding TRB3’s function, it is reported that TRB3 accumulates in response to fasting and inhibits the activation of the kinase Akt in the liver.1,34) TRB3 also stimulates lipolysis by triggering the degradation of acetyl-coenzyme A carboxylase (ACC) in adipose tissue.10)
We thank Tomomi Miyamoto for excellent technical support on the production of transgenic mice. This study was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Ministry of Health, Labour, and Welfare of Japan.