2015 Volume 38 Issue 1 Pages 39-47
2015 Volume 38 Issue 1 Pages 39-47
Cardiac glycosides, which are inhibitors of Na+/K+-ATPase, are classified into cardenolides and bufadienolides. We have recently shown that two cardenolide glycosides, ouabain and odoroside A, inhibit Na+/K+-ATPase, thereby preventing nuclear factor κB-inducible protein expression by blocking Na+-dependent amino acid transport. In this study, we investigated the mechanism of action of cardenolide aglycones in tumor necrosis factor α (TNF-α)-induced gene expression. Ouabagenin, digitoxigenin, and digoxigenin were found to inhibit the TNF-α-induced cell-surface expression of intercellular adhesion molecule-1 (ICAM-1) in human lung carcinoma A549 cells. Those cardenolide aglycones did not inhibit the TNF-α-induced expression of ICAM-1 mRNA, but strongly inhibited the TNF-α-induced expression of ICAM-1 as translation product. The inhibition of the TNF-α-induced ICAM-1 expression by ouabagenin, digitoxigenin, and digoxigenin was significantly reversed by the ectopic expression of ouabain-resistant rat Na+/K+-ATPase α1 isoform. Moreover, knockdown of Na+/K+-ATPase α1 isoform augmented the inhibition of the TNF-α-induced ICAM-1 expression by ouabagenin or ouabain. These results clearly indicate that cardenolide aglycones inhibit the TNF-α-induced ICAM-1 expression at the translation step by blocking Na+/K+-ATPase.
Proinflammatory cytokines, such as tumor necrosis factor α (TNF-α), induce intracellular signaling pathways, one of which triggers the activation of the transcription factor nuclear factor κB (NF-κB).1,2) NF-κB targets a variety of genes essential for inflammation, immunity, and cell survival.3) The cell adhesion molecule, intercellular adhesion molecule-1 (ICAM-1; CD54), is induced mainly by NF-κB.4) ICAM-1 is a cell-surface glycoprotein that belongs to the immunoglobulin superfamily, and serves as a ligand for LFA-1 (CD11a/CD18) and Mac-1 (CD11b/CD18) expressed on leukocytes.5,6) In response to inflammatory mediators, ICAM-1 is upregulated on the vascular endothelial cell surface, and is required for the recruitment and migration of leukocytes to inflamed sites.7) Moreover, it has been shown that immunoglobulin superfamily members, such as ICAM-1, play a role in cancer metastasis.8)
Cardiac glycosides are a family of compounds that function as allosteric inhibitors of Na+/K+-ATPase.9) Cardiac glycosides possess common structures, including a steroid core, an unsaturated lactone ring, and a sugar moiety, and are classified into two groups: cardenolides (five-membered butyrolactone ring) and bufadienolides (six-membered pyrone ring).10,11) It has been reported that the cardenolide glycoside ouabain is able to activate the NF-κB signaling pathway at concentrations that do not inhibit Na+/K+-ATPase activity.12–14) Moreover, it has been reported that cardenolide glycosides, such as oleandrin and digitoxin, inhibit multiple steps in the NF-κB signaling pathway.15–21) We have purified a series of cardenolide monoglycosides, diglycosides, and triglycosides from Nerium oleander, and evaluated their structure–activity relationships of anti-inflammatory and anti-cancer activities.22–25) We initially found that the cardenolide glycoside odoroside A is one of the most active compounds that inhibit TNF-α- or interleukin-1α-induced ICAM-1 expression.22) In contrast to previous reports, we have recently shown that odoroside A and ouabain do not inhibit the NF-κB signaling pathway, but rather prevent the NF-κB-inducible ICAM-1 protein expression by blocking Na+-dependent amino acid transport.26)
Ouabagenin, digitoxigenin, and digoxigenin are the aglycones (sugar-free forms) of three major cardenolide glycosides ouabain, digitoxin, and digoxin, respectively. It has been shown that digitoxigenin and digoxigenin inhibit purified Na+/K+-ATPase with similar efficacy to digitoxin and digoxin, whereas ouabagenin exerts a weaker inhibitory activity than ouabain.27,28) The addition of sugar moieties to the steroid core is not essential for the Na+/K+-ATPase inhibition, but affects the pharmacodynamics and pharmacokinetics of cardenolide glycosides.9,10) Thus far, the biological actions of cardenolide aglycones on the NF-κB-dependent gene expression induced by proinflammatory cytokines have been poorly characterized. We found that ouabagenin, digitoxigenin, and digoxigenin inhibit the TNF-α-induced ICAM-1 expression. In this study, we further investigated the inhibitory mechanisms of those cardenolide aglycones.
Human lung carcinoma A549 cells (JCRB0076) were provided by the National Institute of Biomedical Innovation JCRB Cell Bank (Osaka, Japan). Human hepatocellular carcinoma HepG2 cells (RCB1648) and rat fibroblast 3Y1-B clone 1–6 cells29) (RCB0488) were provided by RIKEN BRC Cell Bank (Tsukuba, Japan). RPMI 1640 medium (Invitrogen, Carlsbad, CA, U.S.A.) and Dulbecco’s modified Eagle’s medium (DMEM) medium (Invitrogen) were supplemented with 10% heat-inactivated fetal calf serum (Nichirei Biosciences, Tokyo, Japan) and the penicillin–streptomycin mixed solution (Nacalai Tesque, Kyoto, Japan). A549 cells and HepG2 cells were maintained in RPMI 1640 medium. 3Y1-B clone 1–6 cells were maintained in DMEM medium.
ReagentsOuabagenin, digitoxigenin, digoxigenin, ouabain, digitoxin, and digoxin were obtained from Sigma-Aldrich (St. Louis, MO, U.S.A.).
Assay for Na+/K+-ATPase ActivityHighly purified Na+/K+-ATPase from porcine kidney (15.6 µmol Pi/min/mg protein at 37°C) was a generous gift from Drs. Yoshikazu Tahara and Yutaro Hayashi (Department of Biochemistry, Kyorin University School of Medicine, Tokyo, Japan).30,31) The ATPase assay was performed as described previously.32)
Cell-Enzyme-Linked Immunosorbent Assay (Cell-ELISA)Cell-surface ICAM-1 expression was measured by the Cell-ELISA as described previously.33,34) Cell-surface ICAM-1 (%) was calculated using the following formula: {(test sample−background)/(sample with TNF-α−background)}×100.
Assay for Cell ViabilityCell viability was evaluated by conducting the crystal violet assay and the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. A549 cells were washed with phosphate-buffered saline (PBS) and then stained with methanol containing 0.2% crystal violet for 15 min. After the cells were extensively washed with water, the crystal violet dye was extracted with methanol. Absorbance at 595 nm was measured. The MTT assay was performed as described previously.33,34) Crystal violet (%) and MTT reduction (%) were calculated using the following formula: {(test sample−background)/(control without TNF-α−background)}×100.
Western BlottingCytoplasmic fractions were prepared and Western blotting was performed as described previously.35,36) Mouse monoclonal antibodies to cyclooxygenase-2 (COX-2) (clone 33; BD Biosciences, Franklin Lakes, NJ, U.S.A.), ICAM-1 (clone 28; BD Biosciences), β-actin (AC-15; Sigma-Aldrich), FLAG (M2; Sigma-Aldrich), and Na+/K+-ATPase α1 (C464.6; Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.) were used for Western blotting. Protein bands were analyzed using an ImageQuant LAS 4000 Mini (GE Healthcare, Piscataway, NJ, U.S.A.).
Real-Time Polymerase Chain Reaction (PCR)Total RNA was prepared with Sepasol-RNA I Super G (Nacalai Tesque) according to the manufacturer’s protocol. Total RNA (1 µg) was reverse-transcribed to cDNA with ReverTra Ace (Toyobo, Osaka, Japan) and oligo(dT)20. cDNA was amplified with a Thermal Cycler Dice® Real Time System Lite (TaKaRa Bio, Otsu, Shiga) using SYBR® Premix Ex Taq™ II (TaKaRa Bio) and the following primers: 5′-CCG AGC TCA AGT GTC TAA AG-3′ (forward primer) and 5′-TGC CAC CAA TAT GGG AAG GC-3′ (reverse primer) for ICAM-1 (369 bp),37) and 5′-ACC ACA GTC CAT GCC ATC AC-3′ (forward primer) and 5′-TCC ACC ACC CTG TTG CTG TA-3′ (reverse primer) for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (452 bp). PCR conditions were as follows: 94°C for 2 min, followed by 40 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s.
TransfectionThe genes encoding human ICAM-1 and rat Na+/K+-ATPase α1 isoform were generated by PCR amplification using the cDNA libraries prepared from TNF-α-stimulated A549 cells and 3Y1-B clone 1–6 cells, respectively. The full-length human ICAM-1 and full-length rat Na+/K+-ATPase α1 were inserted into pCR3-based expression vector containing C-terminal FLAG tag. For transient transfection, A549 cells were transfected with a pCR3 expression vector encoding human ICAM-1 using HilyMax Transfection Reagent (Dojindo Laboratories, Kumamoto, Japan) for 45 h. For stable transfection, A549 cells were transfected with an empty pCR3 expression vector or a pCR3 expression vector encoding rat Na+/K+-ATPase α1 using Lipofectamine® 2000 Transfection Reagent (Invitrogen). The transfected A549 cells were incubated for 2 d in the absence of G418, and then incubated for 12 d in the presence of G418 (1.2 mg/mL). G418-resistant A549 transfectants that did not express rat Na+/K+-ATPase α1 were excluded by incubation with ouabain (1 µM) for 1 d. Two independently isolated A549 clones expressing rat Na+/K+-ATPase α1, designated by RNKAα1 #1 and RNKAα1 #2, were identified by Western blotting using anti-FLAG antibody.
Small Interfering RNA (siRNA) Knockdown of Na+/K+-ATPase α1 IsoformMISSION® siRNA universal negative control #1 and MISSION® siRNA for human Na+/K+-ATPase α1 consisting of 5′-GAG AGA AAG AUACG CCA AAT T-3′ and 5′-UUU GGC GUA UCU UUC UCU CTT-3′ were obtained from Sigma-Aldrich. A549 cells were transfected with either control siRNA or human Na+/K+-ATPase α1 siRNA at the final concentration of 30 nM by using Lipofectamine® RNAiMax Transfection Reagent (Invitrogen) for 48 h.
Statistical AnalysisStatistical significance was assessed by one-way ANOVA followed by the Tukey test for multiple comparisons. A p<0.05 was considered statistically significant.
It has been shown that ouabagenin, digitoxigenin, and digoxigenin inhibit purified Na+/K+-ATPase.27,28) To confirm the specificity of the cardenolide aglycones to Na+/K+-ATPase, highly purified Na+/K+-ATPase from porcine kidney was used for the cell-free ATPase assay. We found that ouabagenin inhibits the ATPase activity at the IC50 value of 1.7 µM with similar efficiency to ouabain (Fig. S1). By contrast, digitoxigenin and digoxigenin inhibited the ATPase activity at the IC50 values of 0.22 µM and 0.87 µM, which were 8.5- and 2.9-fold lower than those of digitoxin and digoxin, respectively (Fig. S1).
Cardenolide Aglycones Inhibit TNF-α-Induced Cell-Surface Expression of ICAM-1ICAM-1 is an NF-κB-responsive gene that is upregulated at the cell surface in response to TNF-α in human lung carcinoma A549 cells.33,34) We have recently shown that ouabain strongly inhibits the TNF-α-induced cell-surface expression of ICAM-1 in A549 cells.26) Ouabagenin was found to inhibit the TNF-α-induced ICAM-1 expression in a dose-dependent manner; its IC50 value was 7.5 µM (Fig. S2), which was 29-fold higher than that of ouabain (Fig. S2). Digitoxigenin and digoxigenin inhibited the TNF-α-induced ICAM-1 expression at the IC50 values of 2.4 µM and 5.1 µM, which were 3-fold and 8-fold higher than those of digitoxin and digoxin (Fig. S2). These data indicate that the cardenolide aglycones inhibit the TNF-α-induced cell-surface expression of ICAM-1, and in contrast to the cell-free ATPase assay, their IC50 values are higher than those of their respective glycoside forms.
To address whether the cardenolide aglycones exert cytotoxic activity against A549 cells, cell viability was evaluated by the crystal violet assay and the MTT assay. Ouabagenin, digitoxigenin, and digoxigenin at concentrations up to 100 µM did not affect or only weakly decreased the crystal violet content during a 7 h incubation (Fig. S3). By contrast, those cardenolide aglycones decreased the MTT reduction by less than 30% under the same conditions (Fig. S4). Ouabain, digitoxin, and digoxin exerted similar effects on the crystal violet content and the MTT reduction at concentrations up to 10 µM (Figs. S3, S4). These data indicate that the cardenolide aglycones do not affect the cell viability, but marginally affect the mitochondrial MTT-reducing activity.
Cardenolide Aglycones Do Not Inhibit TNF-α-Induced Expression of ICAM-1 mRNA, but Selectively Inhibit TNF-α-Induced Expression of ICAM-1 as Translation ProductThe expression of ICAM-1 as translation product was analyzed by Western blotting. TNF-α induced a marked increase in the expression of ICAM-1 protein migrating at around 80 kDa in A549 cells (Fig. 1). The TNF-α-induced expression of ICAM-1 protein was inhibited by ouabagenin at 10–100 µM, and by digitoxigenin and digoxigenin at 1–100 µM (Fig. 1). Ouabain, digitoxin, and digoxin inhibited the TNF-α-induced ICAM-1 protein expression almost completely at concentrations of 1 µM or higher. In addition to ICAM-1, the TNF-α-induced expression of COX-2 protein was inhibited by ouabagenin, digitoxigenin, and digoxigenin (each at 100 µM), and ouabain, digitoxin, and digoxin (each at 10 µM) (Fig. 2A). By contrast, the cardenolide aglycones and the cardenolide glycosides did not inhibit the expression of transfected FLAG-ICAM-1 protein in A549 cells (Fig. 2B) and the constitutive expression of ICAM-1 protein in HepG2 cells (Fig. 2C). These results exclude the possibility that the cardenolide aglycones induce the proteolysis of ICAM-1.

A549 cells were preincubated with various concentrations of ouabagenin, digitoxigenin, digoxigenin, ouabain, digitoxin or digoxin for 1 h, and then incubated with (+) or without (−) TNF-α (2.5 ng/mL) for 6 h in the presence of the compound. Cell lysates were analyzed by Western blotting. The amount of ICAM-1 was normalized to that of β-actin. ICAM-1 protein (%) is shown as the mean±S.E. of three to five independent experiments. * p<0.05 and ** p<0.01, compared with TNF-α (+) compound (−).

(A) A549 cells were preincubated with or without (−) ouabagenin (100 µM), digitoxigenin (100 µM), digoxigenin (100 µM), ouabain (10 µM), digitoxin (10 µM) or digoxin (10 µM) for 1 h, and then incubated with (+) or without (−) TNF-α (2.5 ng/mL) for 6 h in the presence of the compound. Cell lysates were analyzed by Western blotting. The amount of COX-2 was normalized to that of β-actin. COX-2 protein (%) is shown as the mean±S.E. of three independent experiments. ** p<0.01, compared with TNF-α (+) compound (−). (B) A549 cells were transiently transfected with FLAG-tagged ICAM-1 for 45 h. The transfected A549 cells were incubated with or without (−) ouabagenin (100 µM), digitoxigenin (100 µM), digoxigenin (100 µM), ouabain (10 µM), digitoxin (10 µM) or digoxin (10 µM) for 7 h. Cell lysates, including untransfected A549 cells, were analyzed by Western blotting using anti-FLAG antibody. The amount of FLAG-ICAM-1 was normalized to that of β-actin. FLAG-ICAM-1 protein (%) is shown as the mean±S.E. of three independent experiments. (C) HepG2 cells were incubated with or without (−) ouabagenin (100 µM), digitoxigenin (100 µM), digoxigenin (100 µM), ouabain (10 µM), digitoxin (10 µM) or digoxin (10 µM) for 7 h. Cell lysates were analyzed by Western blotting. The amount of ICAM-1 was normalized to that of β-actin. ICAM-1 protein (%) is shown as the mean±S.E. of three independent experiments.
We further investigated the effect of the cardenolide aglycones on ICAM-1 mRNA expression by real-time PCR. TNF-α stimulation markedly induced ICAM-1 mRNA expression to more than 100- to 1000-fold relative to the control within 2 h. The TNF-α-induced ICAM-1 mRNA expression was not inhibited by ouabagenin, digitoxigenin, and digoxigenin (each at 100 µM), and ouabain, digitoxin, and digoxin (each at 10 µM) (Fig. 3). These results indicate that ouabagenin, digitoxigenin, and digoxigenin do not inhibit the TNF-α-induced expression of ICAM-1 mRNA.

A549 cells were preincubated with or without (−) ouabagenin (100 µM), digitoxigenin (100 µM), digoxigenin (100 µM), ouabain (10 µM), digitoxin (10 µM) or digoxin (10 µM) for 1 h, and then incubated with (+) or without (−) TNF-α (2.5 ng/mL) for 2 h in the presence or absence of the compound. Real-time PCR was used to measure the amount of ICAM-1 and GAPDH mRNA. The amount of ICAM-1 mRNA was normalized to that of GAPDH mRNA. ICAM-1 mRNA (%) is shown as the mean±S.E. of four independent experiments. * p<0.05, compared with TNF-α (+) compound (−).
To address whether the cardenolide aglycones inhibit the TNF-α-induced ICAM-1 expression by blocking Na+/K+-ATPase at the cell level, we established A549 cells stably expressing the rat Na+/K+-ATPase α1 isoform. The α1 isoform of rat Na+/K+-ATPase is known to be resistant to ouabain.38) As shown in Fig. 4A, FLAG-tagged rat Na+/K+-ATPase α1 isoform was expressed at similar levels in two independently isolated A549 clones transfected with rat Na+/K+-ATPase α1, designated by RNKAα1 #1 and RNKAα1 #2. Control #1, Control #2, RNKAα1 #1, and RNKAα1 #2 expressed endogenous Na+/K+-ATPase α1 at comparable levels (Fig. 5A). The expression of the transfected rat Na+/K+-ATPase α1 isoform might be very low compared with the expression of endogenous Na+/K+-ATPase α1 isoform, as anti-Na+/K+-ATPase α1 antibody (C464.4) was reactive to human and rat Na+/K+-ATPase α1 isoforms with similar efficacy (data not shown). Ouabain decreased the MTT reduction of Control #1 and Control #2 almost completely at concentrations higher than 0.1 µM during a 48 h incubation (Fig. 4B). By contrast, the MTT reduction was maintained at approximately 70% even when RNKAα1 #1 and RNKAα1 #2 were incubated with ouabain at 10 µM for 48 h (Fig. 4B), indicating that RNKAα1 #1 and RNKAα1 #2 are resistant to ouabain.
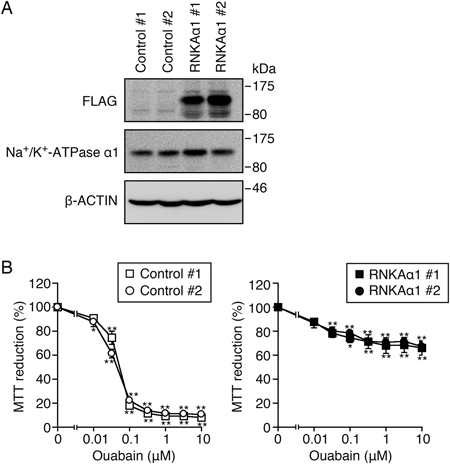
(A) A549 cells were stably transfected with an empty expression vector or an expression vector encoding FLAG-tagged rat Na+/K+-ATPase α1. Cell lysates of two control A549 clones (Control #1 and Control #2) and two rat Na+/K+-ATPase α1-transfected clones (RNKAα1 #1 and RNKAα1 #2) were analyzed by Western blotting. Blots are representative of three independent experiments. (B) Control A549 clones (open squares for Control #1 and open circles for Control #2) and rat Na+/K+-ATPase α1-transfected A549 clones (filled squares for RNKAα1 #1 and filled circles for RNKAα1 #2) were incubated with various concentrations of ouabain for 48 h. MTT reduction (%) is shown as the mean±S.E. of three independent experiments. * p<0.05 and ** p<0.01, compared with ouabain (−).

Control A549 clones (Control #1 and Control #2) and rat Na+/K+-ATPase α1-transfected A549 clones (RNKAα1 #1 and RNKAα1 #2) were preincubated with (+) or without (−) ouabagenin (100 µM), digitoxigenin (100 µM), digoxigenin (100 µM), ouabain (10 µM), digitoxin (10 µM) or digoxin (10 µM) for 1 h, and then incubated with (+) or without (−) TNF-α (2.5 ng/mL) for 6 h in the presence (+) or absence (−) of the compound. Cell-surface ICAM-1 expression was measured by the Cell-ELISA. Cell-surface ICAM-1 (%) is shown as the mean±S.E. of three to four independent experiments. * p<0.05 and ** p<0.01, compared with TNF-α (–) compound (–).
Control A549 clones and rat Na+/K+-ATPase α1-transfected A549 clones were preincubated with cardenolide aglycones (each 100 µM) or cardenolide glycosides (each 10 µM) for 1 h, and then incubated with TNF-α for 6 h in the presence of the compound. In Control #1 and Control #2, the TNF-α-induced ICAM-1 expression was inhibited to the basal levels by ouabagenin, digitoxigenin, digoxigenin, ouabain, digitoxin, and digoxin (Fig. 5). As expected, ouabain inhibited the TNF-α induced ICAM-1 expression by only 10% in RNKAα1 #1 and RNKAα1 #2 (Fig. 5). Moreover, in RNKAα1 #1 and RNKAα1 #2, ICAM-1 expression was significantly induced by TNF-α in the presence of ouabagenin, digitoxigenin, digoxigenin, digitoxin, and digoxin (Fig. 5).
Knockdown of Na+/K+-ATPase α1 Isoform Augments the Inhibition of the TNF-α-Induced Cell-Surface ICAM-1 Expression by Ouabagenin or OuabainTo obtain additional evidence that the cardenolide aglycones block Na+/K+-ATPase at the cell level and thereby inhibit the TNF-α-induced ICAM-1 expression, we performed siRNA knockdown of Na+/K+-ATPase α1 isoform in A549 cells. Transfection of Na+/K+-ATPase α1 siRNA decreased the amount of Na+/K+-ATPase α1 by more than 90% for 48 h (Fig. 6A). Under these conditions, Na+/K+-ATPase α1 knockdown did not affect the crystal violet content, but decreased the MTT reduction by approximately 30% (Figs. 6B, C). These results indicate that Na+/K+-ATPase α1 knockdown does not affect the cell viability, but marginally affects the mitochondrial MTT-reducing activity at least up to 48 h. We found that ouabagenin and ouabain inhibit the TNF-α-induced expression of cell-surface ICAM-1 at lower concentrations in Na+/K+-ATPase α1 knockdown cells than in control cells (Figs. 6D, E). These results indicate that the cardenolide aglycones inhibit the TNF-α-induced ICAM-1 expression by blocking Na+/K+-ATPase.

(A) A549 cells were transfected with control siRNA or human Na+/K+-ATPase α1 (HNKAα1) siRNA for 48 h. Cell lysates were analyzed by Western blotting. The amount of Na+/K+-ATPase α1 was normalized to that of β-actin. Na+/K+-ATPase α1 (%) is shown as the mean±S.E. of three independent experiments. ** p<0.01, compared with control. (B and C) A549 cells were transfected with control siRNA or HNKAα1 siRNA for 48 h. Crystal violet (%) is shown as the mean±S.E. of three independent experiments (B). MTT reduction (%) is shown as the mean±S.E. of three independent experiments (C). ** p<0.01, compared with control. (D and E) A549 cells were transfected with control siRNA (open circles) or HNKAα1 siRNA (filled circles) for 48 h. The siRNA-transfected cells were preincubated with various concentrations of ouabagenin (D) or ouabain (E) for 1 h, and then incubated with TNF-α (2.5 ng/mL) for 6 h in the presence of the compound. Cell-surface ICAM-1 expression was measured by the Cell-ELISA. Cell-surface ICAM-1 (%) is shown as the mean±S.E. of three independent experiments. * p<0.05 and ** p<0.01, compared with TNF-α (+) compound (−).
In this study, we found that three cardenolide aglycones, ouabagenin, digitoxigenin, and digoxigenin, inhibit the TNF-α-induced expression of ICAM-1 at the translation step by blocking Na+/K+-ATPase. By contrast, the cardenolide aglycones did not inhibit the TNF-α-induced expression of ICAM-1 mRNA, indicating that the cardenolide aglycones do not inhibit the NF-κB-dependent signaling pathway. Moreover, in contrast to tunicamycin and brefeldin A that blocked the glycosylation and transport of ICAM-1,36) the cardenolide aglycones did not affect the molecular size of ICAM-1 protein. These data indicate that the cardenolide aglycones do not affect the transport of ICAM-1 to the cell surface. Thus, it seems likely that the cardenolide aglycones act as translation inhibitors in the TNF-α-induced gene expression, except for those capable of inducing the ectodomain shedding of TNF receptor 1 and thereby blocking the TNF-α-induced NF-κB signaling pathway (e.g., glutarimides and triene–ansamycins).39,40)
Na+/K+-ATPase pumps Na+ out and K+ in via hydrolysis of ATP and plays a critical role in the maintenance of the Na+ gradient across the plasma membrane.41) The transport of amino acids across the plasma membrane is mediated by a broad range of transporters, including those coupling the influx of amino acids with the transmembrane gradient of Na+.42) We have previously revealed that odoroside A and ouabain, inhibitors of Na+/K+-ATPase, selectively block Na+-dependent amino acid transport and thereby inhibit the NF-κB-dependent protein expression.26) Consistent with this, it has been reported that cardiac glycosides, including digitoxin, inhibit general protein synthesis by blocking Na+/K+-ATPase.43) In this study, we have shown that three cardenolide aglycones inhibit the TNF-α-induced expression of ICAM-1 protein, but not ICAM-1 mRNA. Moreover, ectopic expression of ouabain-resistant rat Na+/K+-ATPase α1 isoform suppressed the inhibitory effect of ouabagenin, digitoxigenin, and digoxigenin, while knockdown of human Na+/K+-ATPase α1 isoform augmented the inhibitory effect of ouabagenin. These results indicate that the cardenolide aglycones mainly inhibit Na+/K+-ATPase at the cell level in a similar fashion with the cardeonolide glycosides. Thus, we propose a possible mechanism that the cardenolide aglycones inhibit Na+/K+-ATPase and thereby prevent the translation process by blocking Na+-dependent amino acid transport.
The four isoforms (α1, α2, α3, α4) of the Na+/K+-ATPase α subunit exhibit unique tissue distribution.41) The major α1 isoform is found in most tissues and plays a housekeeping role, whereas the α4 isoform is localized in testis.41) It has been reported that the α1, α2, and α3 isoforms are differently expressed in lung cancer cells, including A549 cells.44) However, it seems that the α1 isoform plays a primary role in Na+/K+-ATPase and is more abundant than the other α isoforms in A549 cells, as siRNA targeting the α1 isoform markedly impaired the proliferation and migration of A549 cells for 6 d.44) We have found that knockdown of Na+/K+-ATPase α1 in A549 cells does not affect the cell viability for 48 h, but augments the inhibitory activity of ouabagenin or ouabain on the TNF-α-induced ICAM-1 expression. These results suggest that A549 cells express Na+/K+-ATPase α1 as a major isoform and are capable of resisting a marked reduction of Na+/K+-ATPase α1 level at least up to 48 h. Moreover, the expression of transfected rat Na+/K+-ATPase α1 isoform in ouabain-resistant A549 clones was very low compared with that of endogenous Na+/K+-ATPase α1 isoform. Thus, it seems most likely that a small amount of endogenous Na+/K+-ATPase α1 is sufficient for cell survival and other cellular functions at least for a limited period of time.
It has been reported that ouabain and ouabagenin inhibit the α1β1, α2β1, and α3β1 complexes of human recombinant Na+/K+-ATPase with similar potency, although ouabagenin shows weaker inhibitory activity than ouabain.27) Moreover, ouabagenin and to a lesser extent ouabain inhibit shark Na+/K+-ATPase in a pH-dependent manner.28) Digitoxigenin and digoxigenin inhibit Na+/K+-ATPase to the similar extent as digitoxin and digoxin, and their inhibitory activities are robust to pH.28) In agreement with these studies,27,28) we have confirmed that ouabagenin, digitoxigenin, and digoxigenin directly inhibit the porcine kidney Na+/K+-ATPase activity. However, in contrast to the inhibitory effects on the purified Na+/K+-ATPase activity, ouabagenin, digoxigenin, and digoxigenin at approximately 29-, 3-, and 8-fold higher concentrations than ouabain, digitoxin, and digoxin, respectively, exerted inhibitory activity on the TNF-α-induced ICAM-1 expression. It has been previous reported that ouabagenin has weaker biological activity than ouabain in different cultured cell lines.45,46) Thus, it seems likely that the sugar moiety of the cardenolide glycosides is important for the efficient inhibition of Na+/K+-ATPase expressed on the plasma membrane of the intact cells. In addition, the inhibitory activity of ouabagenin may be influenced by the pH of the culture.
In conclusion, we have shown that cardenolide aglycones inhibit the TNF-α-induced ICAM-1 expression at the translation step by blocking Na+/K+-ATPase. We have recently reported that cardenolide glycosides potently inhibit cellular protein synthesis by blocking Na+-dependent amino acid transport.26) Our studies collectively reveal that both cardenolide aglycones and cardenolide glycosides are able to inhibit translation by blocking Na+/K+-ATPase in the cell. This finding provides invaluable insight into basic and clinical studies based on cardiac glycosides.
We are very grateful to Drs. Yoshikazu Tahara and Yutaro Hayashi for the gift of Na+/K+-ATPase. This work was supported partly by Grants-in-Aid for Scientific Research (KAKENHI) Grant Numbers 22380060 and 25292061 (to T.K.) from the Japan Society for the Promotion of Science.
The authors declare no conflict of interest.
The online version of this article contains supplementary materials.