2015 Volume 38 Issue 4 Pages 571-581
2015 Volume 38 Issue 4 Pages 571-581
Proanthocyanidin is one of the main active compounds found in red jasmine rice. We previously reported that red rice extract could reduce cancer cell invasion. However, the direct effect of proanthocyanidin from red rice on the invasion of cancer cells and the exact molecular mechanism remained unclear. Here, we report for the first time that proanthocyanidin-rich fraction from red rice (PRFR) reduced the migration and invasion of MDA-MB-231 human breast cancer cells. The types of proanthocyanidin in PRFR were identified as procyanidins and prodelphinidins by acid hydrolysis. For cancer cell invasion, degradation of the extracellular matrix (ECM) is required. Treatment of the cells with PRFR reduced the expression of ECM degradation-associated proteins, including matrix metalloproteinase-9 (MMP-9), membrane type-1 matrix metalloproteinase, urokinase plasminogen activator, urokinase plasminogen activator receptor and plasminogen activator-1. Moreover, PRFR also reduced the activity of collagenase and MMP-9. Furthermore, PRFR significantly suppressed the expression of intercellular adhesion molecule-1 and interleukin-6. We also found that PRFR reduced the DNA-binding activity of nuclear factor kappa B (NF-κB), which is the expressed mediator of ECM degradation-associated proteins. These results suggest that proanthocyanidin from red rice mediates MDA-MB-231 breast cancer cell invasion by altering the expression of the invasion-associated proteins, possibly by targeting NF-κB activity.
Breast cancer is a leading cause of mortality among women worldwide including in Thailand. The metastasis of breast cancer to other sites such as the liver, bones, the lungs and lymph nodes is a critical factor contributing to the increased rate of mortality in breast cancer patients.1) The metastasis of cancer cells is a complex process involving cell adhesion, migration and proteolytic degradation of the extracellular matrix (ECM). Degradation of ECM components is an important step in the metastatic process, and it is regulated by the activation of ECM degradation enzymes such as matrix metalloproteinases (MMPs) and urokinase plasminogen activator (uPA).2) Several scientific studies have shown that uPA plays an important role in tumor metastasis and over-expression of uPA in breast cancer is a strong indicator of the poor prognosis.3) The uPA converts the inactive plasminogen into the active serine protease plasmin, which is involved in the degradation of ECM. The uPA system consists of uPA, the uPA receptor (uPAR), plasminogen and its inhibitor, plasminogen inhibitor types 1 and 2 (PAI-1 and -2). The binding of uPA to uPAR not only promotes cancer metastasis but also generates signal transduction that allows enhanced cell survival and migration.3)
MMPs essentially degrade all ECM components surrounding the cancer cells, allowing them to spread from the primary site. Up-regulation of several MMPs such as MMP-2, -3, -9 and membrane type-1 matrix metalloproteinases (MT1-MMP) is generally associated with breast cancer metastasis.4) Therefore, inhibition of expression and activity of ECM proteolytic degradation enzymes is significant for the arrest of cancer cell invasion and metastasis. Transcription factor, nuclear factor kappa B (NF-κB) is a major mediator of inflammation, as well as a regulator of cancer cell development and progression.5) The induction of NF-κB has been shown to regulate the expression of a number of genes whose products are involved in breast cancer cells metastasis. These include ECM degradation enzymes such as MMP-1, MMP-9, MT1-MMP, uPA, cytokine such as interleukin-6 (IL-6) and adhesive molecule, intercellular adhesion molecule 1 (ICAM-1).6)
Many studies have demonstrated that inhibiting NF-κB activity could suppress cancer cell invasion and metastasis by downregulating the expression of their gene production. Therefore, the expression of ECM degradation-associated proteins and their regulatory pathways may represent a useful strategy in the development of anticancer drugs and chemopreventive agents. Proanthocyanidins were identified as dominant antioxidants in red rice varieties.7) Recently, in vitro and in vivo anti-cancer activities of proanthocyanidin in grape seeds have been reported with regard to anti-angiogenesis and cancer invasion.8,9)
In the previous finding, we demonstrated that red rice extract reduced cancer cell invasion via reduced MMP-2, -9 activity and expression.10) However, molecular mechanisms demonstrating how proanthocyanidin obtained from red rice regulating cancer cell invasion has not been investigated and little is known about the mechanism responsible for these effects.
The present study was performed to examine whether proanthocyanidin from red rice inhibits the invasion and motility of breast cancer cells. We examined the effects of proanthocyanidin on ECM degradation-associated proteins, including uPA, uPAR, PAI-1, MMP-9, MT1-MMP, adhesive molecule ICAM-1, cytokine IL-6, and transcription NF-κB in order to understand the molecular mechanism.
Dulbecco’s modified Eagle’s medium (DMEM), penicillin–streptomycin, and trypsin–ethylenediaminetetraacetic acid (EDTA) were purchased from GIBCO-BRL (Grand Island, NY, U.S.A.). Fetal bovine serum (FBS) was purchased from Hyclone (Logan, UT, U.S.A.). Gelatin, β-casein, plasminogen and uPA were obtained from Sigma (St. Louis, MO, U.S.A.). Matrigel was purchased from Becton Dickinson (Bedford, MA, U.S.A.).
Isolation of Proanthocyanidin Rich Fraction from Red RiceWhole grains of red rice (Oryza sativa L.) were harvested from Dok Khamtai cultivar, Phayao Province, Thailand. A voucher specimen number was certified by the herbarium at the Flora of Thailand, Faculty of Pharmacy, Chiang Mai University (voucher specimen No. 023108) which was kept for further reference. The dried and powdered red rice sample (1.0 kg) was extracted with 70% ethanol by being shaken at room temperature for 12 h. After evaporation and lyoplilization, the ethanolic extract was re-dissolved in one liter of hexane–water (1 : 1) and sequentially liquid–liquid partitioned with hexane, dichloromethane and ethyl acetate and the remaining water fraction was further used in isolation of proanthocyanidin.
Proanthocyanidin from red rice was isolated by Sephadex LH20 chromatography as described previously with slight modifications.11) Briefly, the water fraction (5 g) was dissolved in methanol (MeOH) and loaded onto a Sephadex LH-20 Column (20 g hydrated in MeOH and water). The fraction was sequentially eluted with 30% methanol and 70% acetone. Proanthocyanidin in each fraction was determined by vanillin assay. The fractions containing a high concentration of proanthocyanidin were pooled and freeze-dried. This fraction was called proanthocyanidin rich fraction from red rice (PRFR).
The concentration of proanthocyanin in PRFR was determined by acid/butanol assay as described previously with slight modifications.12) Briefly, 10 mg of PRFR was dissolved in 1 mL of butanol–HCl (97.5 : 2.5, v/v) containing 2% w/v NH4Fe(SO4)2·12H2O in 2 N HCl and boiled in hot water for 30 min. The solution was centrifuged at 2500×g for 5 min to get rid of any undissolved material. The absorbance was determined by spectrophotometer at 550 nm and various concentrations of proanthocyanidin from grape seeds were used as the standard curve.
Type Identification of Proanthocyanidin in PRFRTo elucidate the constituent of proanthocyanidin in PRFR, HPLC of the acid hydrolyses of PRFR was performed according to the method described by Oki et al.11) Briefly, 10 mg of PRFR was dissolved in 1 mL of 1.0 M HCl and the mixture was hydrolyzed in boiling water for 30 min. The hydrolysate was re-dissolved in 1.0% (v/v) trifluoroaetic acid (TFA) solution. The identification of the hydrolysate was performed using ultra HPLC H class from Water. An aliquot of the hydrolysate was subjected to a C18-EPS Rocket column (53 mm×7 mm, GRACE) and eluted with isocratic of 0.05% TFA–acetonitrile (87 : 13) for 5 min and detected at 210 nm.
Cell LinesMDA-MB-231 human breast cancer cells, HT1080 human fibrosarcroma, human fibroblast, NIH3T3 mouse fibroblast cells and SKOV-3 human ovarian carcinoma cells were maintained in DMEM, 100 U/mL penicillin and 100 µg/mL streptomycin plus 10% FBS. The cultures were maintained in a humidified incubator with an atmosphere comprised of 95% air and 5% CO2 at 37°C.
Cell Viability AssayMDA-MB-231, HT1080, SKOV-3 (2×103 cells/well) and human skin fibroblast cells (3.5×103 cells/well) were plated in 96-well plates and cultured in DMEM with 10% FBS. After being cultured for 24 h, various concentrations of PRFR (0–200 µg/mL) were added and incubated for 24 or 48 h. At the end of the treatment, 15 µL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (5 mg/mL) were added and it was then incubated for 4 h. The MTT formazan was dissolved with dimethyl sulfoxide (DMSO), and absorbance was measured using a microplate reader at 570 nm with a reference wavelength of 630 nm.
Cell Invasion and Migration AssayThe invasive and migration behavior of MDA-MB-231 cells was tested using a modified Boyden chamber assay as described previously.10) Briefly, polyvinylpyrrolidone-free polycarbonate filters (Millipore) with a pore size of 8 µm were coated with gelatin (0.01%, w/v) for cell migration or with Matrigel (10 µg/ 50 µL) for invasion assay. MDA-MB-231 cells (1×105 cells) were treated with various concentrations of PRFR (0–20 µg/mL) and plated into the upper chamber for 24 h at 37°C in 5% CO2. The medium in the lower chamber contained serum-free culture medium of NIH3T3 cells, which acted as a chemoattractant. Non-migrated cells on the upper surface of the filter were removed and the migrated cells in the lower surface were fixed with methanol and stained with 1% (w/v) toluidine blue. The cells were then dissolved with 20% acetic acid and indirectly quantitated by measuring the absorbance at 570 nm.
Gelatin and Casein ZymographyThe secretion of MMP-9 was analyzed by gelatin zymography as described previously.13) Briefly, MDA-MB-231 cells were treated with various concentrations of PRFR (0–20 µg/mL) for 24 h in DMEM serum-free medium. The culture supernatant was collected and separated by 10% polyacrylamide gels containing 0.1% w/v of gelatin in a non-reducing condition. After electrophoresis, gels were washed twice with 2.5% Triton X-100 for 30 min. The gel was then incubated at 37°C for 18 h in an activating buffer (50 mM Tris–HCl, 200 mM NaCl, 10 mM CaCl2, pH 7.4). Gels were stained with Coomassie Brillant Blue R (0.1% w/v) and destained in 30% ethanol with 10% acetic acid. The activity of MMP-9 was indicated in a clear band against a blue background. The intensity of the digestion bands were quantitated by Bio 1D software (Viber Lourmat).
The inhibitory effect of PRFR on MMP-9 activity was performed by gelatin zymography. The culture supernatant of MDA-MB-231 containing MMP-9 was subjected to 10% polyacrylamide gels containing 0.1% w/v of gelatin as described above. After being washed with Triton X-100, the gel slab was cut into slices corresponding to the lanes and then put in different tanks containing PRFR (0–200 µg/mL) in an activating buffer and incubated at 37°C for 24 h. The MMP-9 activity was quantitated as described above.
The secretion level of uPA from the treated cells was examined by casein-plasminogen zymography.13) The culture supernatant of treated cells was separated by electrophoresis in 10% polyacrylamide gel electrophoresis (PAGE) copolymerized with 1 mg/mL of β-casein and 10 µg/mL human plasminogen under non-reducing conditions. After electrophoresis, gels were washed twice with 2.5% Triton X-100 for 30 min and incubated in activating buffer for 24 h. Gels were stained and destained as described above.
Collagenase Activity AssayFluorometric assay was performed to determine the proteolytic activity of collagenase using EnzChek Gelatinase/Collagenase Assay kit (Molecular Probe).10) Briefly, 1 U/mL of type IV collagenase was mixed with 10 µg/mL of fluorescein-conjugated gelatin (DQ-gelatin) containing PRFR (0–2 µg/mL) in a reaction buffer and dispensed in 96-well microplates. The rate of proteolysis was evaluated by measuring the fluorescence intensity at 3 min interval for 30 min with a fluorometer. The fluorescence values were determined at an excitation wavelength of 485 nm and an emission wavelength of 528 nm. Enzyme activity was estimated by linear regression of the fluorescent intensity recorded during that time.
The Whole Cell Proteins Extraction and Western Blot AnalysisThe whole cell proteins were prepared as previously described.13) Briefly, MDA-MB-231 cells were treated with various concentrations of PRFR for 24 h. The treated cells were washed with ice clod phosphate buffered saline (PBS) and collected by the cell scraper. The cells was lysated with a lysis buffer containing a proteinase inhibitor (50 mM Tris–HCl, 150 mM NaCl, 10 mM, EDTA, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 µg/mL leupeptin, 10 µg/mL aprotinin) for 20 min on ice. The insoluble matter was removed and centrifuged at 12000 rpm for 10 min at 4°C, and the supernatant fraction (whole cell lysate) was collected and protein concentration was determined with the Bradford protein assay. To determine the expression level of the proteins in the whole cell lysate, the protein samples were subjected to 10% sodium dodecyl sulfate (SDS)-PAGE. After electrophoresis, the separated proteins were transferred to a nitrocellulose membrane by electro blotting. The membrane was blocked with 5% nonfat milk in PBS containing 0.5% (v/v) Tween for 1 h and then the membrane was probed with the primary antibodies at 4°C overnight. The membrane was washed and probed with a secondary antibody conjugated with horseradish peroxidase and detected by chemiluminescence (Thermoscientific, Rockford, IL, U.S.A.).
Measurement IL-6 ProductionMDA-MB-231 cells were treated with proanthocyanidin (0–20 µg/mL) for 24 h. After treatment, the culture supernatant was collected and centrifuged at 5000 rpm for 5 min. The level of IL-6 in supernatant was determined using the sandwich enzyme-linked immunosorbent assay (ELISA) kit (Biolegend, San Diego, CA, U.S.A.) according to the manufacturer’s instructions.
DNA Binding Activity of NF-κBThe MDA-MB-231 cells were treated with PRFR (0–20 µg/mL) for 24 h. The treated cells were collected and wash twice with ice cold PBS. The nuclear extraction was performed using the Nucbuster proteins extraction kit (Novagen) according to the manufacturer’s instructions. The DNA-binding activity of NF-κB was evaluated using NF-κB (p65) Transcription Factor Assay Kit (Cayman Company, Ann Arbor, MI, U.S.A.) with slight modifications.13)
The nuclear proteins at 10 µg/mL were added to the well-coated double-stranded DNA (dsDNA) templates carrying NF-κB response elements and incubated at 4°C overnight. Blank wells, positive control (provided with kit) and nonspecific binding samples were also incubated on the plate. After primary (anti-NF-κB) and secondary (goat anti-rabbit horseradish peroxidase (HRP)) antibody treatments, the developing and stopping reagent was added and absorbance was read at 450 nm. The readings for nonspecific binding were subtracted from each treatment. The percent change in activity of each test sample relative to the average of the untreated samples was determined.
Statistical AnalysisAll experiments were performed in triplicate. Quantifications were defined as mean±standard deviation (S.D.) of three independent experiments and expressed as a percentage of the control, which was considered to be 100%. Statistical significances of difference throughout this study were calculated by Student’s t-test. The difference between experimental groups was considered statistically significant when the p value was <0.01 or <0.05.
The proanthocyanidin content in PRFR was analyzed by acid/butanol assay. The total amount of proanthocyanidin in the ethanoic extract and PRFR were 126.75 and 261.38 mg/g extract, respectively. To determine the type of proanthocyanidin in PRFR, the extract was subjected to acid hydrolysis and analyzed by HPLC. Pure gallocatechin (GC), epigallocatechin (EGC), catechin (C), epicatechin (EC) were used as the standard (Fig. 1A). After acid hydrolysis, the peak of gallocatechin, epigallocatechin, catechin and epicatechin were detected by HPLC as shown in Fig. 1B, while there was no HPLC peak that was observed in the non-acid hydrolysis of PRFR (Fig. 1C). This result indicated that proanthocyanidin in PRFR were procyanidin and prodelphinidin.

The effect of PRFR on cell viability of human cancer cells and normal fibroblasts were determined by MTT assay. As shown in Figs. 2A and B, treatment of the cells with various concentrations of PRFR (0–200 µg/mL) was done for 24 and 48 h, no cytotoxicity was observed in the MDA-MB-231, HT1080 and SKOV-3 cells (>80% cell survival). Moreover, at high concentrations of PRFR (200 µg/mL) there was no effect on the viability of normal human fibroblast cells.

Breast cancer cells (MDA-MB-231), ovarian cancer cells (SKOV-3), fibrosarcoma cells (HT1080) and human fibroblast cells were seeded into 96-well plates and treated with PRFR (0–200 µg/mL) for 24 (A) and 48 (B) h The cell viability was determined by MTT assay. The results are expressed as percentages of cell viability relative to the untreated cells and presented as the mean±S.D. of three independent experiments.
To investigate the effect of PRFR on the invasion and migration of MDA-MB-231 human breast cancer cells, a Boyden chamber assay was used. For cell invasion assay, after 24 h of treatment, the cells moved from the upper side of the filter (Matrigel-coated) to the lower one were reduced in a dose dependent manner with IC50 at 7.52±1.42 µg/mL (Fig. 3A). For the cell migration studies, similar experiments were performed using gelatin-coated filters, PRFR inhibited the cell migration dose dependently with IC50 at 10.6±0.59 µg/mL (Fig. 3B).

The upper surfaces of the membrane filters were coated with Matrigel for invasion assay (A) or with gelatin for migration assay (B). MDA-MB-231 1×105 cells were seeded into the upper chamber with different concentrations of PRFR (0–20 µg/mL) and the lower chamber was filled with the culture supernatant of NIH-3T3 cells. After 24 h of incubation, the migrated cells on the lower surface of the filter were determined. The data represent the mean±S.D. of three independent experiments. Sample groups were significantly different from the non-treated group (* p<0.05).
The over-expression and activation of MMP-9 and MT1-MMP was associated with breast cancer metastasis. To investigate possible mechanisms underlying PRFR-mediated anti-invasion property, we examined the effect of PRFR on the activity of MMP-9 and MT1-MMP. As shown in Fig. 4A, the secretion of MMP-9 was suppressed by PRFR in a dose dependent manner with IC50 at 13.5 µg/mL. Moreover, the activity of MMP-9 was reduced to 75% when the enzymes were incubated directly with 200 µg/mL of PRFR (Fig. 4B). In particular, PRFR strongly reduced the activity of collagenase with IC50 at 1.5 µg/mL (Fig. 4C). The results from the Western blot analysis indicated that the expression of MT1-MMP was reduced by PRFR in a dose dependent manner (Fig. 4D).

MDA-MB-231 cells were incubated with PRFR (0–30 µg/mL) for 24 h. The secretion level of MMP-9 was determined with gelatin zymography (A). For MMP-9 activity assay (B), PRFR (0–200 µg/mL) were directly solubilized in the activation buffer. The gel slab was cut into slices corresponding to the lanes and then put in different tanks containing different concentrations of PRFR. The inhibitory effect of PRFR (0–2 µg/mL) on the proteolytic activity of collagenase was measured using a gelatin fluorescent substrate (C). The effect of PRFR (0–15 µg/mL) on the expression of MT1-MMP was determined by Western blot analysis. The data are representative of 3 independent experiments with * p<0.01.
The uPA/uPAR system plays a significant role in the metastasis of many cancer cells, including breast cancer. To evaluate whether the expression level of the uPA/uPAR system was mediated by PRFR, the MDA-MB-231 cells were treated with PRFR for 24 h then, the expressions of uPA, uPAR and PAI-1 were determined by casein zymography and Western blot analysis. As shown in Fig. 5A, PRFR reduced the secretions of uPA in a dose dependent manner. Moreover, the expression levels of uPAR and PAI-1 were decreased dose dependently by PRFR (Fig. 5B).
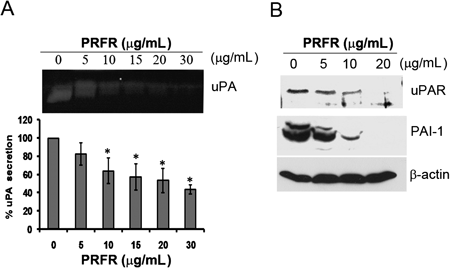
The cells were incubated with PRFR (0–20 µg/mL) for 24 h. The secretion of uPA was determined by casein zymography (A). The expression of uPAR and PAI-1 was determined by Western blot analysis (B). Data from a typical experiment are depicted here, and similar results were obtained in three independent experiments (* p<0.05).
IL-6 is to be pro-inflammatory cytokines that has been reported as poor prognostic factors for the progress of breast cancer patients with regard to tumor metastasis. To investigate the expression level of IL-6 in MDA-MB-231 cells, it was regulated by PRFR. The level of IL-6 in the cell culture supernatant was determined by ELISA. As shown in Fig. 6A, the level of IL-6 was reduced to 58% by treatment with 15 µg/mL of PRFR. ICAM-1 have been shown to be highly expressive in breast cancer and associated with cancer cell adhesion and metastasis. In order to determine the effect of PRFR on the expression of ICAM-1, MDA-MB-231 cells were treated with PRFR for 24 h and the level of ICAM-1 was determined by Western blot analysis. Treatment with 20 µg/mL of PRFR resulted in a decrease in the expression level of ICAM-1 in the cells (Fig. 6B).
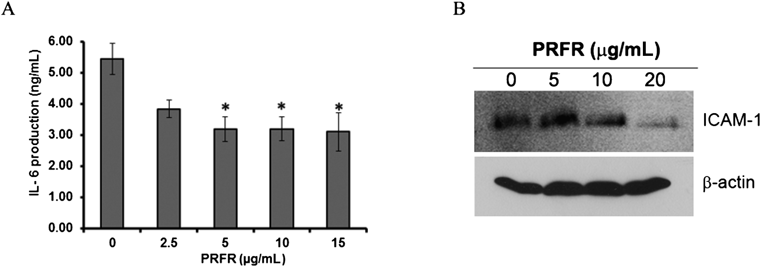
MDA-MB-231 cells were incubated with various concentrations of PRFR for 24 h in DMEM serum free medium. The culture supernatant was collected to determine the level of IL-6(A). The cells were treated with lysis buffer to determine the expression level of ICAM-1 by Western blot (B). The data are representative of 3 independent experiments with * p<0.01.
NF-κB plays a key role in the expression of MMPs, uPA, uPAR, cytokines and ICAM-1, which were all involved in cancer metastasis. Therefore, we analyzed the NF-κB DNA binding activity of MDA-MB-231 cells to determine the effect of PRFR at the transcription level of those proteins. As shown in Fig. 7, treatment of the cells with various concentrations of PRFR for 24 h reduced the DNA binding activity of NF-κB in a dose dependent manner with IC50 at 4.2 µg/mL.

The cells were incubated with various concentrations of PRFR for 24 h. Nucleus extracts were prepared and used to analyze NF-κB DNA-binding activity by ELISA. The data represent the mean±S.D. of three independent experiments. Sample groups were significantly different from the untreated group (* p<0.05).
Proanthocyanidins are oligomer and polymer of flavanol-3-ol unit which found in various fruits, vegetables and cereals including grape seeds, pine barks, berries, and red rice. Proanthocyanidin can be classified into several classes depending on the hydroxylation of the constitutive units and the linkages between them.14) The common units are (epi)catechins and (epi)gallocatechins, leading to procyanidin and prodelphinidin structures, respectively. The bonding between monomeric units in proanthocyanidin are most commonly linked via C-4 to C-8 or C-6 (type B proanthocyanidin). Our results demonstrated that monomeric units of proanthocyanidin in acid hydrolysis of PRFR were catechins, epicatechin, gallocatechin, and epigallocatechin. These results revealed that the proanthocyanidin in PRFR was the procyanidin (catechin and/or epicatechin) and prodelphinidin (epigallocatechin and/or gallocatechin) types as the chemical structure shown in Figs. 8A and B, respectively. This finding was similar to the report of Bordiga et al.15) which demonstrated that four flavan-3-ol monomer were found in red rice. They also identified the major type of proanthocyanidin in red rice as procyanidin B-1 (epicatechin-(4β to 8)-catechin) with the mean degree of polymerization (DP) was 4.2. Whereas, low amount of prodelphinidin was also observed. In the agreement with the reported from Min et al.,16) the structural character of proanthocyanidin in IITA119 red rice are procyanidin and consist mainly in oligomer of 5–8 monomer (40%), while the polymer (DP>10) accounted for 29%. Oki et al. indicated that the proanthocyanidin in the extracts of red-hull rice was the procyanidin type, which possess a DP ranging from 1 to 38 monomer.11) Whereas, the proanthocyanidin in grape seed is procyanidin type and represents major oligomer with DP4 (60%).17) Based on Sephadex LH gel filtration column chromatography, the PRFR was co-eluted at the same fractions (fraction numbers 27 to 29) as the proanthocyanidin from grape seed extract as shown in the supplement data 1. Therefore, we propose that the majority of proanthocyanidin in the PRFR are the oligomer with the same DP of proanthocyanidin found in the grape seed extract as the structure was proposed in Fig. 8C. In addition, proanthocyanidin has been widely distributed in plant food and present high structural diversity, which make the differences in biological and physical properties. The proanthocyanidins in pine bark mainly comprise of procyanidin type B together with small amount of prodelphinidin and contain in oligomer with chain length between two and pentadecamer.18) On the other hand, the major type of proanthocyanidin in lingonberry is proanthocyanidin type A and mainly present in pentamer and heptamer.19) Bioavailability of proanthocyanidin is largely influenced by their degree of polymerization; while monomer, dimer and trimer are readily absorbed by small intestine, polymer and oligomer need to be biotransformed by acid conditions in the stomach and/or by micro flora in the colon.20) Therefore, the intact and decomposed products of proanthocyanidin may exert heath beneficial effects throughout the digestive tract after absorption.

Procyanidins (A) and prodelphinidins (B), and structure representative of red rice proanthocyanidin; tetramer of procyanidin type B1 (C).
In the recent years, the biological properties of proanthocyanidin from red rice have been studied extensively, including for anti-oxidant, anti-inflammation and anticancer properties.21) However, there has been no direct evidence to show that proanthocyanidin extract obtained from red rice exerts an effect on cancer cell metastasis. In this study, we investigated the anti-metastasis property of proanthocyanidin from red rice on MDA-MB-231 breast cancer cells and explored its molecular mechanism. Our results indicate for the first time that non-cytotoxic doses of PRFR significantly reduced MDA-MB-231 cancer cell invasion and migration even at a very low concentration level. The PRFR inhibitory concentration (IC50=7.5 µg/mL) is in the same concentration range as the grape seed extract reported by Dinicola et al.9) In addition, our previous report has shown that the proanthocyanidin from grape seed extract inhibited MDA-MB-231 cell invasion.22) On the other hand, inhibition of MDA-MB-231 cells invasion by evening primrose extract was at higher concentration (IC50 ca. 60 µM). The major proanthocyanidin in evening primrose extract are in dimer and trimer form.23) This additional evidences confirm that proanthocyanidin in PRFR are the oligomers which might have a similar structure to the proanthocyanidin in the grape seed extract.
The MMPs play an important role in tissue remodeling associated with various pathological processes such as angiogenesis, cirrhosis, arthritis, and cancer metastasis.24) However, it appears that MMP-2 and MMP-9 play a key role in cancer cell invasion and metastasis that can degrade type IV collagen, one of the major components of basement membrane.25) MT1-MMP is a transmembrane metalloprotease that plays a major role in the extracellular matrix remodeling directly by degrading several of its components and indirectly by activating pro-MMP-2.26) An over-expression of collagenase, MMP-9 and MT1-MMP have all been associated with a high potential of breast cancer cell metastasis.27) Our results from gelatin zymography showed that PRFR reduced the MMP-9 secretion and activity. Moreover, PRFR strongly inhibited collagenase activity when DQ-gelatin was used as a substrate. On the other hand, PRFR strongly reduced the expression of MT1-MMP in MDA-MB-231 cells. This result was similar to previous findings which have stated that proanthocyanidin derived from Japanese quince fruit inhibited MMP-2 and MMP-9 enzyme activity.28) In addition, treatment of the blueberry proanthocyanidin-enriched fraction also reduced MMP-2 and -9 activities in DU145 human prostate cancer cells.29) However, other MMPs such as MMP-1 and MMP-3 are thought to be important in rheumatoid arthritis and osteoarthritis.30) while MDA-MB-231 cells do not secret MMP-2.
Many reports have shown the correlation of uPA and uPAR in breast cancer cells metastasis in terms of decreasing or knocking down the level of these proteins, potentially reducing cancer cell invasion.3,31) Our results demonstrated that a low concentration level of PRFR successfully decreased the secretion of uPA and the expression of uPAR. Although contradictory findings have been reported for the levels of PAI-1 that are involved in cancer cell metastasis. These conflicting findings may relate to the fact that PAI-1 are multifunctional proteins and their effect on cancer progression may be both context- and concentration-dependent. A pro-tumorigenic action of PAI-1 can be reasoned from numerous clinical studies implying high serum levels of PAI-1 as a predictor of poor prognostic for patients with ovarian, gastric and breast cancers.32,33) Besides PAI-1, PAI-2 can also bind to uPA but does so slower than PAI-1. High secretion of PAI-2 was observed in placenta during pregnancy. Moreover, several reports have been indicated that PAI-2 encourages cancer cell growth by inhibiting apoptosis and the high expression of PAI-2 was found in cervical, oral, colon and lung cancers.34,35)
PAI-1 can potentially influence cells motility at several levels by competing with uPAR and intergrins to bind vitronectin which promotes the cells ability to detach from ECM.36) Furthermore, a down regulation in the expression of PAI-1 in MDA-MB-231 cells was associated with an inhibition of the cell invasion.37) Our results demonstrated that PRFR strongly reduced PAI-1 expression, suggesting that PRFR decreases MDA-MB-231 cell invasion, at least in part, via reduced uPA, uPAR and PAI-1 expression levels. These results correlate with previous reports that have stated that proanthocyanidin from grape seeds reduced uPA, uPAR and PAI-1 expression.38) Taking the results together, it is suggested that PRFR reduced cancer cell invasion, at least in part, due to the inhibited activity and expression of ECM degradation proteins.
Cytokines, such as tumor necrosis factor-alpha (TNF)-α, IL-6 and IL-8, produced from tumors and/or activated immune cells have altered tumor cell behavior through complex mechanisms. IL-6 expression in the breast tumor microenvironment can impact the host’s immune defense mechanisms in terms of tumor cell growth, proliferation, differentiation and angiogenesis. It also induces metastasis through the up-regulation of the cancer cells’ adhesion and invasion capacity.39) The signal transduction of IL-6 involves the activation of several oncogenic pathways such as the signal transducer and activator of transcription-3 (STAT-3) and mitogen-activated protein kinase (MAPK) signaling pathways. Recent data have demonstrated that IL-6 induced MDA-MB-231 cell invasion via the STAT-3 signaling pathway.40) Moreover, inhibition of IL-6 signaling has been shown to attenuate tumor growth and angiogenesis and metastatic events in breast cancer patients.
Our results show that PRFR inhibits the level of IL-6 in MDA-MB-231 cells, which suggests that PRFR reduced the key functional cytokines that promote the cell migration and invasion ability. Moreover, the effects of PRFR on the expression of ICAM-1 in MDA-MB-231 cells were studied. Cancer cell adhesion to endothelial cells is an essential process during cancer metastasis. Several adhesive molecules, such as ICAM-1, VCAM-1 and E-selectin, have been identified as being responsible for the endothelial adhesion of cancer cells.41) While overexpression of ICAM-1 has been found in several cancer cells including those in breast cancer subjects.42) Therefore, agents that repress ICAM-1 expression in breast cancer cells and block the adhesion of cancer cells might be therapeutic targets for the repression of the metastasis potential of cancer cells. Our result showed that PRFR reduced the expression of ICAM-1 in the MDA-MB-231 breast cancer cells. Similar to our findings, Kalfin et al. demonstrated that proanthocyanidin from grapeseeds reduced the level of ICAM-1 in the sclerosis patient.43)
The NF-κB transcription factor is constitutively activated in many types of cancer including in breast cancer cells. The NF-κB signaling pathway is known to regulate the expression of various genes involved in the process of tumor metastasis. Several studies have shown that the inactivation of the NF-κB DNA binding activity in MDA-MB-231 cells inhibited the expression of many downstream target genes involved in tumor metastasis, such as MMP-9, MT1-MMP, uPA, uPAR, vascular endothelial growth factor (VEGF), COX-2, ICAM-1 and IL-6.44) Interestingly, our results demonstrated that the treatment of MDA-MB-231 cells with PRFR strongly inhibited the NF-κB DNA binding activity. These results demonstrated a strong link among NF-kB, MMP-9, MT1-MMP, uPA, uPAR, IL-6 and ICAM-1 expression and are also in agreement with recent reports on the inhibition of NF-κB by proanthocyanidin in prostate cancer cells.45)
Overall, our data suggest that PRFR could be a powerful candidate in the development of preventive agents against breast cancer metastasis. On the other hand, PRFR may act as a promising adjuvant therapy combined with modern anti-cancer drugs. PRFR is likely to inhibit the metastasis cascade affecting MMP, uPA, IL-6 and ICAM-1. Further investigations in clinical studies will be required to assess the potential of PRFR in the treatment of cancer.
This work was supported by the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission, the Agricultural Research Development Agency (Public Organization) (ARDA) and the Faculty of Medicine, Chiang Mai University, Thailand.
The authors declare no conflict of interest.
The online version of this article contains supplementary materials.