2015 Volume 38 Issue 4 Pages 611-617
2015 Volume 38 Issue 4 Pages 611-617
Metal responsive element (MRE)-binding transcription factor-1 (MTF-1) is a zinc finger (ZF) transcription factor that plays a key role in heavy metal homeostasis by regulating relevant genes in response to metals. MTF-1 is known to be activated by heavy metals such as Zn and Cd, but the mechanism of activation remains unclear. In the present study, Cys and His residues of human MTF-1 (hMTF-1), some of which may be involved in interaction with metals or with each other, were screened for their contribution to Zn-dependent transcription. To avoid poor induction ratios of previous transfection assays, we re-examined experimental conditions to establish an assay able to correctly detect Zn-responsive transcription. Using this assay, a series of Cys and/or His substitution mutants were analyzed over the entire hMTF-1 molecule. In five out of the six ZFs (ZF1 to ZF5), Cys mutations that disrupt the ZF structure abolished response to Zn. Of these, ZF5 was shown for the first time to be essential for Zn-responsive transcription, despite it being unnecessary for Zn-induced DNA binding. These results indicate that Zn activation of hMTF-1 involves an additional process besides induction of DNA binding activity. Our assay also confirmed the importance of Cys in the acidic activation domain, as well as those in the C-terminal Cys cluster, implicated in transcription in other studies. The identified Cys residues might contribute to metal response of hMTF-1 through direct metal binding and/or intramolecular interactions, analysis of which will be helpful in understanding the mechanism of metal response.
Metal responsive element (MRE)-binding transcription factor-1 (MTF-1) is a transcription factor that plays a central role in controlling cellular heavy metals.1–3) In mammalian cells, this protein is known to regulate genes encoding metallothioneins (MTs) involved in homeostasis of essential metals as well as protection against toxic metals,4) ZnT1 involved in Zn transport,5) and a number of other proteins.6,7) MTF-1 is activated when cells are exposed to heavy metals such as Zn and Cd,8,9) consequently inducing transcription of the relevant genes by binding to a specific regulatory DNA sequence, MRE.10,11) The molecular mechanism of MTF-1 activation has been enthusiastically investigated because of its physiological and toxicological importance, but has not been fully understood yet. In protein-DNA binding experiments using cell extracts, Zn added in vitro induced binding of MTF-1 to MRE,12,13) implying a mechanism of metal induction.
MTF-1, consisting of 753 amino acids in humans and 675 amino acids in mice,14) contains six consecutive Cys2–His2 zinc fingers (ZFs) in the N-terminal half of the molecule and three transcriptional activation domains (acidic, Pro-rich, and Ser/Thr-rich domains) in the C-terminal half.8) In studies of deletion mutants, only those retaining the ZF domain can bind to DNA,8,11) and an isolated ZF domain specifically recognizes and binds to MRE,15) suggesting that the ZF domain functions as a DNA-binding domain, as with conventional Cys2–His2 ZFs. The ZF domain by itself appears insufficient for metal-dependent DNA-binding, and coexistence of other regions of the MTF-1 molecule is required.11,16) These findings remind us a mechanism analogous to those reported for yeast transcription factors, in which more than one functional domain is involved in metal regulation.17) Putative regulatory roles have been proposed for some ZFs of hMTF-1 based on physicochemical analyses of an isolated ZF domain,15,18) but no conclusive evidence has been obtained yet. Also, some regions outside the ZF domain are required for metal-dependent transcription,19,20) but the mechanisms by which they function and their relationships with the ZF domain remain unclear.
In the present study, we intended to determine the amino acid residues essential for metal-induced transcriptional activity of hMTF-1 by a comprehensive analysis of point mutants. Prior to this analysis, we made efforts to improve the quality of a transfection assay, which had been less reliable in correctly estimating metal response. Using an improved assay, hMTF-1 mutants with substitutions at Cys and/or His residues, some of which might possibly be involved in interaction with metals or with each other, were analyzed.
Immortalized MTF-1(−/−) mouse embryonic fibroblasts (dko7),8) a generous gift from Dr. Walter Schaffner, were cultured in Dulbecco-modified minimum essential medium plus 10% fetal calf serum.
PlasmidsA reporter plasmid p(MREa)4MTprCAT has a minimal promoter of the human metallothionein-IIA (hMT-IIA) gene fused to the reporter chloramphenicol acetyltransferase (CAT) gene, with four tandem copies of the hMT-IIA MREa sequence inserted immediately upstream of the promoter.11) pMT-CAT (described in our previous report21) as pUC-MTCA T) contains the hMT-IIA promoter (up to −767) fused to the CAT gene. pRSVL, a reference plasmid, expresses the firefly luciferase gene under the control of the Rous sarcoma virus long terminal repeat promoter.22) pCI-MTF-1 (described in our previous report11) as pCI-hMTF-1b) contains hMTF-1 cDNA inserted downstream of the cytomegalovirus immediate early enhancer/promoter in the expression vector pCI (Promega, U.S.A.).
Expression plasmids of hMTF-1 mutants each with a Cys-to-Tyr substitution in one of the six ZFs have been described previously.11) Expression plasmids of other MTF-1 mutants were prepared using a PrimeSTAR Mutagenesis Basal Kit (Takara Bio, Shiga, Japan), according to the manufacturer’s instructions. All introduced mutations were confirmed by DNA sequencing. For convenience, the mutated Cys and His residues are indicated by a one-letter code, and are numbered in order from the N-terminus; e.g., C4 and H15 represent the fourth Cys and fifteenth His residues from the N-terminus, respectively. In principle, Cys residues were substituted by Tyr or Ser, and His residues were substituted by Asn. Some mutants have more than one substitution at closely positioned target amino acids. For H15 and H16/H17 within ZF5 which constitute extra His residues in Cys2–His2 ZF, HisAsp and HisHis sequences were converted to GlnGlu and TyrSer which are the sequences found at the corresponding positions in ZF2, respectively, to avoid disruption of the finger structure. Details concerning individual amino acid substitutions are indicated in the figures.
Northern Blottingdko7 cells were plated in 10-cm dishes at 2.5×105 cells/10 mL medium with streptomycin (100 µg/mL) and penicillin (100 units/mL), and cultured for 24 h. The cells were transfected with pMT-CAT (8 µg) and pCI-MTF-1 (1 µg) using the standard calcium phosphate method. After 24 h, the cells were incubated with or without 100 µM ZnSO4 for 6 h before harvest. Extracted RNA samples were analyzed by Northern blotting with digoxigenin (DIG)-labeled RNA probes, as described previously.23) RNA probes for mouse MT-I (mMT-I) and CAT transcripts were prepared as described,23) using an RsaI fragment of mMT-I cDNA containing the entire coding region and a PvuII/XbaI fragment of the pCAT3 Basic plasmid (Promega) covering the C-terminal 182 amino acids of CAT, respectively.
Transient Transfection AssayTransient transfection assays were carried out according to the standard calcium phosphate transfection protocol (Figs. 1, 2b), or using a transfection reagent FuGENE HD (Roche; Figs. 2c, 3–5). CAT levels in cell lysates were analyzed using CAT-ELISA (Roche). In experiments using the reference plasmid pRSVL, luciferase activity was measured as described previously,22,24) and relative reporter activity, i.e., CAT level normalized relative to luciferase activity, is indicated unless otherwise stated.
For experiments using calcium phosphate transfection, details of experimental conditions are indicated in the figure legends. Transfection using the FuGENE HD reagent was carried out according to the manufacturer’s instructions. dko7 cells were plated in 35-mm dishes at 2×105 cells/2 mL medium, and cultured for 24 h. The cells were transfected with pMT-CAT (0.4 µg), pRSVL (0.2 µg) and pCI-MTF-1 or a hMTF-1 mutant expression plasmid (0.02 µg). The total amount of DNA to be transfected was adjusted to 1 µg by adding pUC19 DNA. An optimal FuGENE HD reagent : DNA ratio of 3 : 1 was adopted. After 4 h the cells were incubated with or without 100 µM ZnSO4 for 4 h before harvest.
Concerning the endogenous MT gene known to be regulated by MTF-1, expression is restricted to very low levels unless cells are exposed to inducers.25) By contrast, in a transfection assay with overexpressed MTF-1 and an MRE-containing reporter vector, high basal levels of reporter activity were observed, making it difficult to correctly assess metal response.11) Prior to the analysis of hMTF-1 mutants, we first investigated the cause of such high basal activity. As a possible explanation, overexpressed MTF-1 might neutralize a putative negative regulator that suppresses the activity of MTF-1 in the absence of inducers.26) To verify this hypothesis, we examined whether metal response depends on the levels of recombinant hMTF-1 expressed in MTF-1(−/−) dko7 cells (Fig. 1). Expression of an MRE-regulated CAT reporter gene showed high basal activity at 25 ng pCI-MTF-1 and was almost unresponsive to Zn, consistent with our previous results obtained using HeLa cells.11) If this had been due to a negative regulator, dilution of the input hMTF-1 would have rescued response to Zn. However, Zn response was never recovered, even when the quantity of pCI-MTF-1 was lowered to 0.2 ng, at which only a near-background level of activity was detected. During this study, we noted that another reporter plasmid pMT-CAT containing the intact hMT-IIA gene upstream sequence (up to −767) fused to the CAT gene showed higher CAT expression levels and improved Zn response, probably due to a synergistic effect of the MRE’s arranged in the promoter.22) Accordingly, this plasmid was used for further analysis.
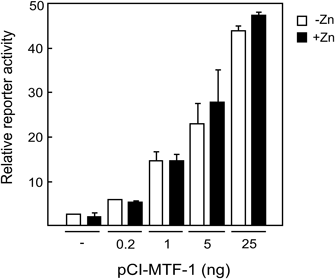
dko7 cells were plated at 105 cells/6-cm dish and cultured for 3 d. The cells were transfected with p(MREa)4MTprCAT (3.2 µg), pRSVL (0.8 µg) and the indicated amounts of pCI-MTF-1 using a standard calcium phosphate transfection method (the total DNA amount was adjusted to 4.4 µg/plate with pUC19 DNA). After 45 h the cells were treated with (filled bars) or without (open bars) 100 µM ZnSO4 for 4 h, and relative reporter activity was determined as described in Materials and Methods. Averages of data from assays carried out in duplicate are shown with errors.
To see if the observed high basal expression occurs at the transcriptional level, we next determined CAT-mRNA levels in pMT-CAT-transfected dko7 cells cultured with or without Zn (Fig. 2a). In contrast to the high basal CAT protein levels observed in Fig. 1, CAT-mRNA expression levels were low in the absence of Zn, and markedly increased with Zn treatment (Fig. 2a, lower panel). Endogenous mMT-I gene expression was also induced by Zn, but uninduced levels were more strictly limited (Fig. 2a, upper panel). These results suggested that the relatively high basal expression of CAT-mRNA might lead to the accumulation of CAT protein with a longer half life. To verify this idea, we determined CAT protein levels before and after induction by Zn (Fig. 2b). Control cells incubated for 6 h without Zn (Fig. 2b, open bars) showed high CAT levels, at approximately 70% of Zn-induced levels (filled bars). Nearly the same levels of CAT were observed for cells harvested without 6 h incubation (hatched bars), indicating that the high basal levels were due to CAT accumulation before Zn induction. These results suggested that optimal conditions for detecting response to Zn could be achieved by minimizing CAT accumulation. To test this, we used a transfection protocol using a FuGENE HD reagent that enables more sensitive detection of reporter activity. In fact, shortening the period between transfection and Zn induction to 4 h successfully lowered basal levels with an induction ratio of 9-fold (Fig. 2c) as compared with 1.5-fold in Fig. 2b. Zn induction for 6 h as in Fig. 2b gave a similar induction ratio (data not shown), and a more convenient 4-h induction protocol used in Fig. 2c was adopted in the analysis of hMTF-1 mutants described below.
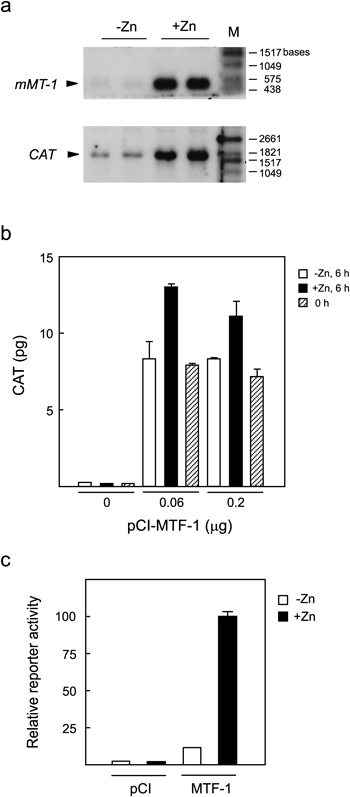
a: Relatively high basal expression of the CAT reporter gene. dko7 cells were transfected with pMT-CAT and pCI-MTF-1 using the calcium phosphate method and cultured for 24 h, followed by incubation with or without 100 µM ZnSO4 for 6 h further. Extracted RNAs were electrophoresed in formaldehyde–1% agarose gel and transcripts coding for endogenous mMT-1 (upper panel) and CAT (lower panel) were detected with DIG-labeled RNA probes. Data for duplicate plates are shown. M indicates DIG-labeled RNA molecular weight markers (Roche). b: CAT protein accumulates prior to Zn induction. dko7 cells were plated at 5×104 cells/35-mm dish and cultured for 24 h. The cells were transfected with pMT-CAT (1.6 µg) and the indicated amount of pCI-MTF-1 by calcium phosphate transfection (total DNA amount was adjusted to 2 µg by adding pUC19). After 24 h, the cells were harvested (hatched bars) or incubated for 6 h further with (filled bars) or without (open bars) 100 µM ZnSO4 followed by harvest, and CAT levels in cell lysates were determined. Average CAT values from assays performed in duplicate are shown with errors. c: Improved assay with a shorter Zn induction time. dko7 cells were transfected with pMT-CAT and pRSVL together with pCI-MTF-1 or an equimolar amount of the empty expression vector pCI using FuGENE HD reagent. After 4 h the cells were incubated with or without 100 µM ZnSO4 for a further 4 h, and relative reporter activity was determined as described in Materials and Methods. Data from two independent experiments each carried out in duplicate were normalized relative to the value for MTF-1-transfected, Zn-treated cells (taken as 100), and averages with standard errors are shown.
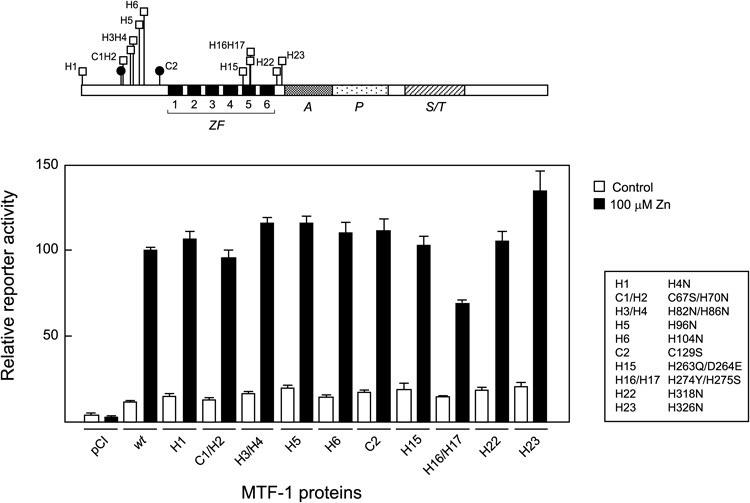
The interactions of amino acid residues with metals or with each other might possibly be involved in metal regulation of MTF-1, and Cys and His residues could act as interfaces for such interactions. Hence we constructed a library of mutants each with an amino acid substitution(s) at Cys and/or His residues in the hMTF-1 molecule, and analyzed them for Zn-responsive transcription using the improved assay described above. First, hMTF-1 mutants with defective ZFs were analyzed. Each mutant had a substitution of the second Cys in one of the six Cys2–His2 ZFs, representing loss-of-function mutants of these ZFs. Although these mutants had been analyzed in our previous work,11) HeLa cells used in that study had endogenous MTF-1 activity, making the results unclear together with the usually observed high basal activity. Hence these mutants were analyzed again using the improved assay (Fig. 3). Mutants of ZF1 to ZF4 (substitutions at C4, C6, C8 and C10, respectively) almost completely lost basal as well as Zn-induced activity. Mutation of ZF5 (substitution at C12) also reduced both basal and Zn-induced activity, but to a lesser extent. Only mutation of ZF6 (substitution at C14) showed no effect. In this experiment, as well as those in Figs. 4 and 5, it was confirmed that the loss of activity was not due to decreased protein expression levels by immunoblotting (data not shown).

Locations of the mutations in the hMTF-1 molecule are indicated at the top of the figure. ZF, zinc finger domain (each ZF is indicated by a black box and numbered); A, acidic domain; P, Pro-rich domain; S/T, Ser/Thr-rich domain. Mutated Cys residues are indicated by one letter code numbered in the order from the N-terminus, e.g. C4 representing the fourth Cys. Detailed information for the mutants is shown on the right of the figure. dko7 cells were transfected with pMT-CAT, pRSVL and an expression plasmid of hMTF-1 variant with a mutation in one of the ZFs. Four hours later the cells were incubated with (filled bar) or without 100 µM ZnSO4 (open bar) for 4 h further, and relative reporter activity was determined as described in Materials and Methods. All experiments included the cells transfected with wild type (wt) hMTF-1 and an empty expression vector pCI, to enable direct comparison between experiments. Data from three independent experiments each carried out in duplicate were normalized relative to the value for wt hMTF-1-transfected and Zn-induced cells (taken as 100), and averages with standard errors are shown.
Mutants with substitutions at Cys and/or His residues within the N-terminal half of the hMTF-1 molecule were assayed for Zn-responsive transcription and the results are indicated as in Fig. 3. Mutated Cys and His residues are indicated by a one-letter code numbered in order from the N-terminus (e.g., C2 and H5 represent the second Cys and fifth His, respectively).
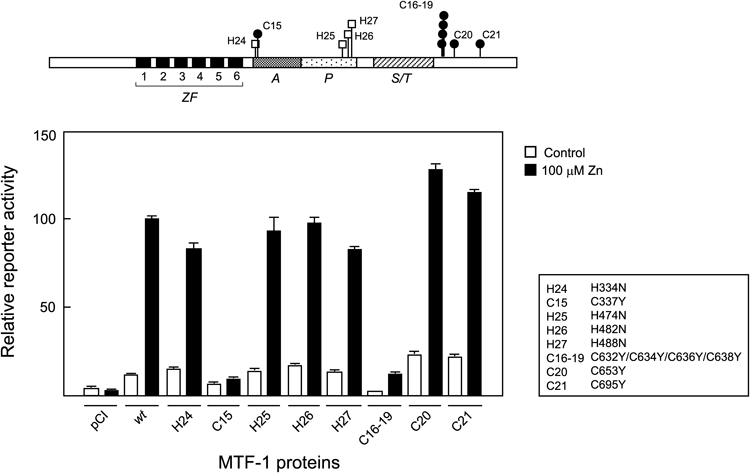
Mutants with amino acid substitutions within the C-terminal half of the hMTF-1 molecule were assayed for Zn-responsive transcription as in Fig. 3 and the results are indicated as in Fig. 4.
Next, mutants with substitutions at Cys and/or His residues other than those conserved in the Cys2–His2 ZFs were analyzed. Results for the residues located in the N-terminal half of hMTF-1 are shown in Fig. 4. In general, each mutant had a single amino acid substitution, but in some cases where the relevant residues were located in close proximity to one another (e.g. C1 and H2), more than one substitution was introduced in a single mutant plasmid. None of the mutations examined had a large effect. Only in the H16/H17 double mutant, Zn-induced activity was reduced by about 30%.
Cys and His residues located in the C-terminal half of hMTF-1 were also analyzed (Fig. 5). Impaired Zn response was observed in two mutants. One was the C15 mutant with a substitution of the Cys residue located at the N-terminus of the acidic activation domain. In this mutant, Zn-induced activity was almost completely abolished, and basal activity was also decreased by half. The other mutant had substitutions at C16–19 which constitutes a Cys cluster located C-terminal to the Ser/Thr-rich region. Zn-induced activity was reduced to near the basal level of wild type (wt) hMTF-1, and basal activity was almost completely abolished.
Analyses of MTF-1 mutants in previous studies have been carried out under a variety of experimental conditions different in species, cell types, measuring objects and analytical methods. Such experimental divergence may be responsible, at least in part, for the often observed discrepancies between study results. In this context, analysis of MTF-1 mutants under a unified and reliable condition seemed necessary. Although transient transfection assays provide a quite useful way to assess metal-dependent transcriptional activity, there has previously been a problem concerning low metal induction ratios with high uninduced basal activity.9,11,26) Increased metal response has been reported in cells cultured in serum-free medium,19) but apparent metal-induced reporter activity could reflect not only transcriptional activation but also subcellular localization, since MTF-1 tends to reside in the cytoplasm under serum-free conditions and heavy metals can induce its translocation to the nucleus.27) In the present study, we investigated assay conditions that enable detection of metal-induced transcription in conventional serum-containing medium. High basal expression was found to be due to the accumulation of CAT protein before metal induction, resulting from elevated basal mRNA expression. Together with the observation of limited basal expression of the hMT-IIA gene,25) endogenous chromatin-packaged MT genes appear to be more strictly controlled in the absence of metals than a reporter gene on a plasmid. Consequently, an assay appropriate for evaluating metal responsive transcription with low basal activity was achieved by adjusting conditions in order to minimize CAT accumulation. This improvement also made the assay more convenient, which can be completed in a shorter time.
Amino Acid Residues Required for Zn-Responsive TranscriptionTranscriptional activity of yeast heavy metal-responsive transcription factors such as Mac1 and Zap1 is considered to be regulated by intramolecular interactions modulated by Cu or Zn ions.17) Mammalian MTF-1 might also be regulated by a similar mechanism involving multiple functional domains, based on observations that the loss of only a part of the MTF-1 molecule severely affects transcriptional activity,8) and that Zn-dependent DNA binding of MTF-1 requires not only the DNA binding domain but also additional parts of the molecule.11,16) From a functional screening of Cys and His residues potentially involved in metal regulation, several Cys residues were identified to be necessary for Zn-responsive transcription (Figs. 3–5). These include Cys residues located in five ZFs (C4, C6, C8, C10 and C12, respectively), the acidic activation domain (C15), and the C-terminal Cys cluster (C16–19).
To date, differential roles of the six ZFs of MTF-1 have been discussed. N-Terminal four ZFs, ZF1 to ZF4, have a strong affinity to Zn and are postulated to be the canonical Cys2–His2 ZFs involved in DNA-binding.15,18) Mutations in these ZFs prevent DNA binding and consequently impair transcription.8,11,18) Consistent with these observations, the results shown in Fig. 3 confirm that mutations in ZFs 1–4 abolish basal and metal-induced transcription. It has been suggested that ZF1 and the linker between ZF1 and ZF2 are involved in metal regulation of mMTF-1,9,28) while their roles have not been confirmed in hMTF-1.3,11,29)
In contrast to the N-terminal ZFs, ZF5 and ZF6 show low Zn affinity and have been assumed to play regulatory roles.15,18) However, their deletion mutants9) as well as point mutants11) show a phenotype similar to wt MTF-1 both in heavy metal-induced MRE binding and transcriptional activity. Nevertheless, the assay developed in the present study demonstrates for the first time that a mutation in ZF5 impairs Zn-responsive transcription. It seems likely that the effect of this mutation on Zn response was masked by the high basal levels of our previous assay.11) It should be noted that ZF5 is essential for Zn-induced transcription in spite of the fact that it is unnecessary for Zn-dependent DNA binding.11) This indicates that Zn activation of hMTF-1 requires another distinct process in addition to induction of DNA binding. ZF5 mutation did not completely abolish both basal and Zn-induced activity, showing characteristics dissimilar from mutations in ZF1 to ZF4.
The C15 residue located within the acidic activation domain and overlapping the nuclear export signal27) was also essential for response to Zn. The C15 mutant shown in Fig. 5 has a Cys to Tyr substitution, the latter being the amino acid at the corresponding position in mMTF-1. It has been reported that the Zn-dependent activity of a chimeric transcription factor, consisting of a Gal4 DNA-binding domain and an hMTF-1 acidic activation domain, is reduced by the conversion to this mouse-type amino acid.20) Our data demonstrate that this mutation impairs the Zn response of intact hMTF-1 as well. Another mutant with a Cys to Ser substitution at the same position showed a similar phenotype (data not shown). Leu residues located near this position in hMTF-127) and mMTF-130) were also reported to be important for Zn-induced transcription. Our assay also determined the C-terminal Cys cluster to be the third site essential for response to Zn. This region has been reported to be required for Zn-dependent transcription but unnecessary for DNA-binding and subcellular localization,19) and is likely to function as a dimerization interface.31) Our results confirm the importance of these Cys residues. In addition, mutation of residues H16/H17 resulted in partial loss of activity (Fig. 4). These residues may be involved in base recognition.1)
Future Studies for Understanding the Mechanism of Zn RegulationComprehensive screening of hMTF-1 mutants in our present study identified Cys and His residues required for Zn-responsive transcription, and also confirmed that other Cys and His residues are not essential. Analysis of residues that may be important for metal binding and intra- or intermolecular interactions would provide insights into the mechanism of metal regulation. In particular, ZF5, with its low Zn affinity allowing reversible association with Zn under physiological conditions, is likely to play a role in metal regulation.15,18) The affinity of an isolated ZF domain to MRE was not markedly affected by saturation of the low-affinity ZFs with Zn ions,15) leading us to envisage an interaction between ZF5 and another site outside the ZF domain. Residues C15 located near ZF5 and C16–19 might be candidates for a possible interaction interface. Investigation into interactions between these residues would provide clues to enable understanding of metal regulation. It remains to be determined whether residues other than Cys and His contribute to metal regulation.
The authors declare no conflict of interest.