2016 Volume 39 Issue 1 Pages 68-77
2016 Volume 39 Issue 1 Pages 68-77
Approximately 180 t/km2 of Asian dust particles are estimated to fall annually on Beijing, China, and there is significant concern about the influence of microbes transported by Asian dust events on human health and downwind ecosystems. In this study, we collected Asian dust particles in Beijing, and analyzed the bacterial communities on these particles by culture-independent methods. Bacterial cells on Asian dust particles were visualized first by laser scanning microscopy, which demonstrated that Asian dust particles carry bacterial cells to Beijing. Bacterial abundance, as determined by quantitative polymerase chain reaction (PCR), was 108 to 109 cells/g, a value about 10 times higher than that in Asian dust source soils. Inter-seasonal variability of bacterial community structures among Asian dust samples, as compared by terminal restriction fragment length polymorphism (T-RFLP), was low during the Asian dust season. Several viable bacteria, including intestinal bacteria, were found in Asian dust samples by denaturing gradient gel electrophoresis (DGGE). Clone library analysis targeting 16S ribosomal RNA (rRNA) gene sequences demonstrated that bacterial phylogenetic diversity was high in the dust samples, and most of these were environmental bacteria distributed in soil and air. The dominant species in the clone library was Segetibacter aerophilus (Bacteroidetes), which was first isolated from an Asian dust sample collected in Korea. Our results also indicate the possibility of a change in the bacterial community structure during transportation and increases in desiccation-tolerant bacteria such as Firmicutes.
Aeolian dust primarily occurs in arid and semi-arid regions. When a wind-sand stream occurs, dust particles can be lifted and transported over long distances by air currents. Aeolian dust particles are thought to be carriers of microbes, and abundance and community composition of microbes transported with aeolian dusts have been reported to clarify their possible impacts on public health and ecosystems.1–5) Major aeolian dust events arise from the Sahara and Sahel deserts (African dust), Australian deserts (Australian dust), and the Taklamakan Desert, Gobi Desert and Loess Plateau (Asian dust). It is well known that Asian desert dust particles are transported for long distances,6) even reaching the North American Continent (more than 15000 km away).7,8) Asian dust particles can sometimes be transported globally in 13 d9) and have been identified in ice and snow cores of Greenland10) and the French Alps.11)
In China, Asian dust has caused disasters as sand storms.12) Approximately 180 t/km2 of Asian dust particles are estimated to fall annually on Beijing, China.13) There are significant concerns regarding the health effects of Asian dust14–18) as well as the influence of microbes transported by Asian dust events on plant, animal and human diseases and on downwind ecosystems.19–22) To estimate the risk of bacteria transported by Asian dust events on human health and downwind ecosystems, the abundance and community structure of bacteria carried on Asian dust particles should be accurately determined. It is well-known that most (usually more than 90%) bacteria in the natural environment are difficult to culture under conventional conditions23) and thus culture-independent approaches are required for analysis of bacterial dynamics in atmospheric environments.24,25)
In this study, we collected Asian dust particles in Beijing, and visualized bacterial cells on Asian dust particles by laser scanning microscopy to directly demonstrate that Asian dust particles carry bacterial cells. We determined bacterial abundance by quantitative polymerase chain reaction (PCR) targeting the eubacterial 16S ribosomal RNA (rRNA) gene and compared bacterial community profiles by terminal restriction fragment length polymorphism (T-RFLP) among Asian dust samples to investigate inter-seasonal variability of bacterial community structure during the Asian dust season. We also analyzed bacterial communities before and after culturing with four media to estimate viable bacterial genera and species by denaturing gradient gel electrophoresis (DGGE) based on bacterial 16S rRNA gene sequences. Finally we analyzed bacterial community structure by clone library analysis targeting 16S rRNA gene sequences to determine the dominant bacteria in Asian dust samples collected in Beijing.
Asian dust samples that fell on the top of a building (ca. 10 m in height) were collected at the China Agricultural University in Beijing (Figs. 1A, B). Dust particles that fell into a sterilized stainless-steel bucket over a 24 h period were collected. Twelve samples were obtained on different days in the Asian dust season between 20 March and 11 May, 2010. When severe Asian dust events occur in Beijing, dust particles could cover a car (Fig. 1C), and more dust particles were collected within 24 h on 20 March and 10 May than on the other sampling days.
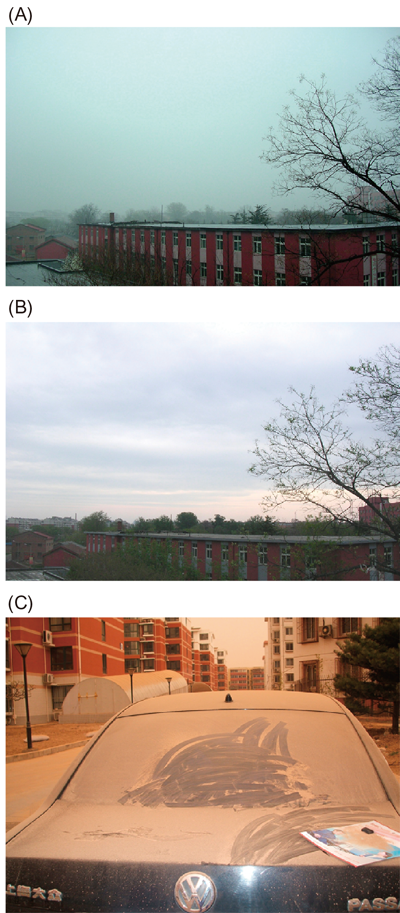
When a severe Asian dust event occurred, a car was covered with dust particles within one night (C).
Ten milligram of the dust samples was suspended in phosphate-buffered saline (PBS) (pH 7.2) containing 4% (w/v) paraformaldehyde and fixed at 4°C for 16 h. After fixation, dust suspensions were filtered through sterilized 0.4 µm-pore size polycarbonate membrane filters (K040A025A; Toyo Roshi Kaisha, Tokyo, Japan) and rinsed twice with particle-free water. The filters were then dehydrated in an ethanol series (50, 80 and 100% ethanol for 3 min each) and dried under vacuum. The filters were stained for 10 min with nucleic acid-staining dye, SYBR Green I (Life Technologies, Carlsbad, CA, U.S.A.; 1/10000-dilution of the supplied product) containing 2% Tween 20. After washing with particle-free water (3 times, 10 min each), the filters were mounted in immersion oil for observation by laser scanning microscopy (TCS-SP5; Leica Microsystems, Wetzlar, Germany). The microspectrophotometer of the laser scanning microscope was used to discriminate microbial cells from dust particles.26) All particles were observed under blue excitation (wavelength 480 nm) according to the optimized protocol26) and particles with green fluorescence (wavelength 500–540 nm) were detected as microbial cells; those with yellow to red fluorescence (wavelength 550–680 nm) were detected as non-biological particles such as soil particles.26)
Direct DNA ExtractionTotal nucleic acids were extracted from 0.5 g of each Asian dust samples using a FastDNA Spin Kit for soil (MP Biomedicals, Santa Ana, CA, U.S.A.) according to the manufacturer’s instructions. The DNA suspension was purified using a Wizard DNA clean-up system (Promega, Fitchburg, WI, U.S.A.) according to the manufacturer’s instructions.
Quantitative Real-Time PCRBacterial abundance was quantified by real-time PCR targeting 16S rRNA genes with a LightCycler (Roche Diagnostics, Basel, Switzerland). Real-time PCR was performed with universal primer sets EUB f933 (5′-GCA CAA GCG GTG GAG CAT GTG G-3′) and EUB r1387 (5′-GCC CGG GAA CGT ATT CAC CG-3′)27) according to the procedure reported by Nishimura et al.28) To determine the recovery rate of DNA during extraction, known amounts of DNA fragment of the luciferase gene (luc) were inoculated into the samples before DNA extraction as an internal standard and quantified after DNA extraction.28) The DNA recovery rate was calculated by comparing the copy number of the inoculated luc gene before and after DNA extraction. The copy number of the 16S rRNA gene quantified by real-time PCR was calibrated based on the DNA recovery rate.
T-RFLP AnalysisFor the T-RFLP analysis, 16S rRNA gene sequences of the bacterial population were amplified by PCR using the universal primers Cy5-labeled-8f and EUB926r.29) PCR for T-RFLP analysis was performed according to the procedure reported by Yamaguchi et al.30) PCR products were purified and concentrated using MiniElute PCR purification Kit (QIAGEN, Hilden, Germany). The purified PCR products were then digested with MspI (TaKaRa Bio, Shiga, Japan) for 3 h at 37°C. DNA was precipitated with ethanol and the DNA pellet was then suspended in CEQ Sample Loading Solution (Beckman Coulter, Brea, CA, U.S.A.). CEQ DNA Size Standard-600 (Beckman Coulter) was added as an internal standard and fluorescence labeled terminal restriction fragments (T-RFs) were size separated by capillary electrophoresis using the CEQ8000 (Beckman Coulter). T-RFs shorter than 90 bp were excluded from the analysis to avoid uncertainties associated with fragment size determination as reported by Hodges and Olson.31) Most peaks derived from dust samples were larger than 90 bp. The measured peak profiles were analyzed using the database MiCA3 (http://mica.ibest.uidaho.edu/rundigest.php).
DGGE AnalysisTo identify active bacterial species, DGGE analysis was performed according to the procedure reported by Iwamoto et al.27) on both directly extracted bacterial DNA and bacterial DNA extracted from cultured Asian dust samples. To culture bacterial cells in Asian dust samples collected on 20 March and 10 May, 10 mg of each dust sample was inoculated into 9 mL of four media; standard medium (5 mg/mL peptone, 2.5 mg/mL yeast extract, and 1 mg/mL glucose), 1% standard medium, 0.05% yeast extract, and R2A medium (Nihon Pharmaceutical, Osaka, Japan). Samples were incubated at 25°C for 7 d with slow shaking, and then bacterial DNA was extracted and purified using the Wizard Genomic DNA Purification Kit (Promega) according to the manufacturer’s instructions. DGGE and sequencing of DGGE fragments were performed according to the procedure reported by Yamaguchi et al.32)
Random CloningThe 16S rRNA gene library was constructed from the Asian dust sample collected on 20 March. Nearly full-length 16S rRNA gene sequences of the bacterial domain were amplified by PCR using the universal primers 8f (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492r (5′-TAC CTT GTT ACG ACT T-3′), according to the procedure reported by Yamaguchi et al.26) 16S rRNA gene fragments of more than 150 randomly selected clones were sequenced at Fasmac (Kanagawa, Japan). No clones were obtained when DNA suspension was not added. Sequences were analyzed with the ribosomal database project33) and clustered as operational taxonomic units (OTUs) based on a 97% cutoff. Rarefaction curves were constructed with Distance-Based OTU and Richness (DOTUR) at 85% similarity.34)
Microbial cells on Asian dust particles collected in Beijing were visualized by laser scanning microscopy using an optimized protocol26) following fluorescent nucleic acid staining with SYBR Green I (Fig. 2). We confirmed that microbial cells were attached to big dust particles (>5 µm) and several microbial cells were attached to one of these ‘big’ particles (Fig. 2). No microbial cells were attached to small particles (<5 µm). These data directly demonstrate that Asian dust particles carry microbial cells to Beijing.

(A) Green fluorescence from bacterial cells under blue excitation. (B) Red fluorescence from Asian dust particles under blue excitation. (C) Composite image.
Bacterial abundance on Asian dust particles was determined by quantitative real-time PCR targeting the 16S rRNA gene with universal primer sets. Asian dust samples collected in Beijing during the spring Asian dust season were shown in Table 1. Abundance of bacteria on dust particles was between 1×108 and 2×109 (average: 8×108) cells/g. The source region of Asian dust particles during this Asian dust season was determined to be the Gobi Desert by HYSPLIT back trajectory analysis (http://ready.arl.noaa.gov/HYSPLIT_traj.php). We have determined bacterial abundance in the Gobi Desert to be between 5×106 and 4×108 (average: 9×107) cells/g sand.35) The number of bacterial cells transported to Beijing with Asian dust particles was about 10 times higher than those in the dust source regions. Bacterial abundance in general soil environments ranges from 1010–1011 cells/g.36) Soil particles which are lifted up from the ground by strong wind can be mixed with Asian dust particles during transportation37) from the dust source region to Beijing, and thus bacterial abundance of Asian dust particles could increase.
| Samples | 16S rRNA gene (copies/g) | Estimated bacterial number* (cells/g) |
|---|---|---|
| 20 Mar. | 1×109 | 2×108 |
| 21 Mar. | 4×109 | 8×108 |
| 23 Mar. | 1×109 | 2×108 |
| 24 Mar. | 8×108 | 2×108 |
| 27 Mar. | 9×109 | 2×109 |
| 4 Apr. | 4×109 | 8×108 |
| 9 Apr. | 8×109 | 2×109 |
| 10 Apr. | 4×109 | 8×108 |
| 29 Apr. | 1×109 | 2×108 |
| 7 May | 6×108 | 1×108 |
| 10 May | 1×1010 | 2×109 |
| 11 May | 4×109 | 8×108 |
* 16S rRNA gene copy number: 5 copies/cell.
T-RFLP targeting the bacterial 16S rRNA gene as a universal genetic marker can be widely used for characterization of bacterial community composition.29) T-RFLP is semi-quantitative and has a high reproducibility. T-RFLP has been applied for community analysis of bacteria in natural environments (e.g., lakes,38) ocean,39) soil40) and air24)), because this method is suitable for evaluation of temporal and spatial changes in the targeted microbial communities. T-RFLP analysis was therefore performed in this study with DNA fragments obtained from 12 Asian dust samples, to examine the similarity of bacterial communities41) among Asian dust samples. The T-RFLP profiles were similar among the 12 samples, and two dominant OTUs with lengths of 150 and 275 bp were detected in all Asian dust samples (Fig. 3) while some specific OTUs (445 and 495 bp) were detected in a few samples collected on 29 April and 7 May. These measured peak profiles were analyzed by the database MiCA3. Bacterial species producing the 150 bp fragment under the T-RFLP conditions used in this study were Azospirillum brasilense (Proteobacteria; nitrogen-fixing bacteria), Azospirillum zeae (Proteobacteria; nitrogen-fixing bacteria), Beijerinckia indica (Proteobacteria; nitrogen-fixing bacteria), Methylobacterium nodulans (Proteobacteria; nitrogen-fixing bacteria), Methylobacterium radiotolerans (Proteobacteria; radiation tolerating bacteria), Paenibacillus polymyxa (Firmicutes; nitrogen-fixing bacteria), Sphingobium japonicum (Proteobacteria; soil bacteria), Sphingomonas melonis (Proteobacteria; plant pathogenic bacteria). In addition, Bacillus sp. (spore-forming bacteria frequently isolated from soil), uncultured Bacteroidetes bacterium and Rubellimicrobium sp. (atmospheric bacteria) also produce the 150 bp fragment under the T-RFLP conditions used in this study. Bacteria producing the 275, 445 and 495 bp length fragments were Corynebacterium pseudotuberculosis (Actinobacteria; frequently isolated from soil and pathogen of livestock42)), Polynucleobacter necessaries (Proteobacteria; free-living freshwater bacteria) and Salmonella enterica (Proteobacteria; intestinal bacteria), respectively. These results are summarized that nitrogen-fixing bacteria frequently found in natural environments, especially in soil, were detected and some pathogenic and intestinal bacteria were also found by T-RFLP analysis.

Arrows indicate 150, 275, 445 and 495 bp, respectively.
DGGE was used to determine the viable bacterial community in Asian dust samples collected in Beijing. The sequences of 52 bands on DGGE gels were determined (19 bands from uncultured Asian dust samples (Fig. 4A) and 33 bands from cultured Asian dust samples (Fig. 4B). Phylogenetically diverse bacteria were confirmed to exist in the Asian dust samples (Tables 2, 3).

(A) Uncultured dust samples. (B) Dust samples cultured with a) standard medium, b) 1% standard medium, c) 0.05% yeast extract, and d) R2A medium.
| Band | Closely related species | Number of matched bases (similarity) | |
|---|---|---|---|
| 20 Mar. | A | Hymenobacter sp. | 444/456 (97%) |
| B | Paracoccus sp. | 401/431 (93%) | |
| C | Methylobacteriaceae bacterium | 451/454 (99%) | |
| D | Shinella sp. | 442/457 (96%) | |
| E | Rubellimicrobium aerolatum | 416/435 (95%) | |
| F | Leptolyngbya sp. | 440/455 (96%) | |
| G | Planococcus sp. | 457/457 (100%) | |
| H | Rubellimicrobium sp. | 313/324 (96%) | |
| I | Rubellimicrobium sp. | 428/433 (98%) | |
| J | Skermanella sp. | 453/458 (98%) | |
| 10 May | K | Adhaeribacter aquaticus | 440/459 (95%) |
| L | Niastella koreensis | 438/457 (95%) | |
| M | Methylobacteriaceae bacterium | 457/459 (99%) | |
| N | Kaistia sp. | 438/459 (95%) | |
| O | Shinella sp. | 440/457 (96%) | |
| P | Hymenobacter sp. | 453/456 (99%) | |
| Q | Rubellimicrobium aerolatum | 412/430 (95%) | |
| R | Rubellimicrobium sp. | 428/433 (98%) | |
| S | Skermanella sp. | 453/458 (98%) |
| Band | Closely related species | Number of matched bases (similarity) | |
|---|---|---|---|
| 20 Mar. | 1 | Clostridium aciditolerans | 444/452 (98%) |
| 2 | Clostridium botulinum | 425/445 (95%) | |
| 3 | Clostridium sp. | 450/452 (99%) | |
| 4 | Clostridium sp. | 433/452 (95%) | |
| 5 | Salmonella enterica | 451/456 (98%) | |
| 6 | Salmonella enterica | 453/456 (99%) | |
| 7 | Erwinia sp. | 450/456 (98%) | |
| 8 | Erwinia sp. | 455/456 (99%) | |
| 9 | Bifissio spartinae | 445/457 (97%) | |
| 10 | Adhaeribacter aquaticus | 452/456 (99%) | |
| 11 | Magnetospirillum bellicus | 455/458 (99%) | |
| 12 | Sphingobacterium sp. | 439/457 (96%) | |
| 13 | Telmatospirillum siberiense | 437/459 (95%) | |
| 14 | Bacteroidetes bacterium | 413/455 (90%) | |
| 15 | Bacteroidetes bacterium | 413/456 (90%) | |
| 16 | Methylophaga thalassica | 405/455 (89%) | |
| 17 | Dechlorospirillum sp. | 441/455 (96%) | |
| 10 May | 18 | Clostridium algidixylanolyticum | 343/345 (99%) |
| 19 | Clostridium sp. | 451/453 (99%) | |
| 20 | Clostridiaceae bacterium | 449/453 (99%) | |
| 21 | Ensifer sp. | 459/459 (100%) | |
| 22 | Pseudomonas sp. | 457/457 (100%) | |
| 23 | Cupriavidus necator | 450/452 (99%) | |
| 24 | Flavosolibacter ginsengiter | 454/455 (99%) | |
| 25 | Flavosolibacter sp. | 447/457 (97%) | |
| 26 | Deinococcus sp. | 452/459 (98%) | |
| 27 | Microvirga sp. | 459/460 (99%) | |
| 28 | Pedobacter ginsengisoli | 453/456 (99%) | |
| 29 | Flavobacterium sp. | 445/454 (98%) | |
| 30 | Bacteroidetes bacterium | 449/456 (98%) | |
| 31 | Clostridium magnum | 440/453 (97%) | |
| 32 | Bacillus sp. | 455/455 (100%) | |
| 33 | Stenotrophomonas sp. | 456/458 (99%) |
In uncultured bacterial population (Table 2), soil bacteria such as Hymenobacter sp., Paracoccus sp., Methylobacteriaceae, Shinella sp., Planococcus sp., Skermanella sp., Niastella koreensis, Kaistia sp. were found as well as atmospheric bacteria (Rubellimicrobium spp.) and aquatic bacteria (Adhaeribacter aquaticus). As shown in Table 2, Hymenobacter sp., Methylobacteriaceae, Shinella sp., Rubellimicrobium spp. and Skermanella sp. were found in the both Asian dust samples collected in March and May.
In cultured Asian dust samples (Table 3), soil bacteria such as Clostridium spp., Ensifer sp., Flavosolibacter spp., Microvirga sp., Pedobacter ginsengisoli, Bacillus sp. and Stenotrophomonas sp. were detected. As shown in Table 3, Clostridium spp. and uncultured Bacteroidetes bacterium were found in the both Asian dust samples collected in March and May. In addition to these bacteria found in natural environments, intestinal bacteria (Salmonella enterica and Erwinia sp.) were detected. Salmonella enterica were also detected in the T-RFLP analysis as previously described. These intestinal bacteria are sometimes found in fecal compost, and they may be recent dust contaminants from soil environments with fecal compost between Beijing and the dust source area, because these intestinal bacteria are thought to lose their activities by UV exposure and desiccation during transportation. Magnetospirillum bellicus and Dechlorospirillum sp. were also detected and these bacteria were described in DDBJ as found in activated sludge used in wastewater treatment plants. There are several wastewater treatment plants around Beijing (http://www.gcus.jp/report/wholeReport/document/pdf/gesuidouten2010_1-03.pdf) and these data may therefore support the explanation for the increase in bacterial abundance during transportation from the dust source area to Beijing, as previously mentioned in relation to results in Table 1.
Methylobacteriaceae, Rubellimicrobium sp., Salmonella enterica, uncultured Bacteroidetes bacterium, and Bacillus sp. were detected in the both T-RFLP and DGGE analysis.
Phylogenetic Analysis of Asian Dust Bacterial CommunityThe bacterial community on Asian dust particles was further analyzed using a clone library targeting the bacterial 16S rRNA gene. As mentioned above, T-RFLP analysis indicated that bacterial communities were similar among 12 Asian dust samples, and phylogenetic analysis was therefore performed on the Asian dust sample collected on 20 March. As shown in Fig. 5, major phyla identified from the dust sample were the members of Bacteroidetes (26%), Actinobacteria (19%), Proteobacteria (18%) and Firmicutes (14%), and this result was similar to the results reported in previous studies on phylogenetic analysis of bacterial composition in aeolian dust.1,2) Jeon et al. collected Asian dust samples in Korea and reported that dominant bacterial phyla were the members of Firmicutes, Actinobacteria and Proteobacteria.43) The obtained clone library also supported the results obtained by T-RFLP analysis with the MiCA3 database and DGGE analysis. Actually, uncultured Bacteroidetes bacteria, Rubellimicrobium spp. (Proteobacteria; soil bacteria) and Bacillus spp. (Firmicutes; spore-forming bacteria frequently isolated from soil) were detected in the all analyses (T-RFLP, DGGE and clone library). In the clone library (Table 4), one clone of Bacteroidetes (unclassified to species level by DDBJ), two clones of Rubellimicrobium (one was R. mesophilum44) and the other was R. roseum45)) and four clones of Bacillus spp. (all were unclassified to species level) were obtained.

| Accession No. | Phylum | Family | Closest described species |
|---|---|---|---|
| Accession No. | Phylum | Family | Closest described species |
| LC026921 | Acidobacteria | Acidobacteriaceae | Acidobacteria bacterium Ac_11_E3 |
| LC026871 | Acidobacteria | Acidobacteriaceae | Acidobacteria bacterium KBS 96 |
| LC026841 | Acidobacteria | Acidobacteriaceae | Acidobacteria bacterium LP6 |
| LC026877 | Acidobacteria | Acidobacteriaceae | Acidobacteria bacterium LWQ4 |
| LC026880 | Acidobacteria | Acidobacteriaceae | Acidobacteria bacterium LWQ4 |
| LC026907 | Acidobacteria | Acidobacteriaceae | Acidobacteria bacterium WWH8 |
| LC026844 | Acidobacteria | Blastocatellaceae | Aridibacter kavangonensis |
| LC026845 | Acidobacteria | Blastocatellaceae | Aridibacter kavangonensis |
| LC026933 | Acidobacteria | Blastocatellaceae | Blastocatella fastidiosa |
| LC026932 | Actinobacteria | Pseudonocardiaceae | Actinomycetospora straminea |
| LC026815 | Actinobacteria | Micrococcaceae | Arthrobacter globiformis |
| LC026857 | Actinobacteria | Micrococcaceae | Arthrobacter phenanthrenivorans |
| LC026899 | Actinobacteria | Micrococcaceae | Arthrobacter phenanthrenivorans |
| LC026838 | Actinobacteria | Cellulomonadaceae | Cellulomonas aerilata |
| LC026931 | Actinobacteria | Cellulomonadaceae | Cellulomonas aerilata |
| LC026903 | Actinobacteria | Microbacteriaceae | Curtobacterium sp. BM-8J |
| LC026840 | Actinobacteria | Nocardioidaceae | Friedmanniella lacustris |
| LC026897 | Actinobacteria | Nocardioidaceae | Friedmanniella lacustris |
| LC026924 | Actinobacteria | Nocardioidaceae | Friedmanniella lacustris |
| LC026925 | Actinobacteria | Nocardioidaceae | Friedmanniella lacustris |
| LC026856 | Actinobacteria | Nocardioidaceae | Friedmanniella luteola |
| LC026864 | Actinobacteria | Nocardioidaceae | Friedmanniella luteola |
| LC026891 | Actinobacteria | Nocardioidaceae | Friedmanniella luteola |
| LC026818 | Actinobacteria | Nocardioidaceae | Friedmanniella sp. I10A-01996 |
| LC026843 | Actinobacteria | Nocardioidaceae | Friedmanniella sp. I10A-01996 |
| LC026866 | Actinobacteria | Nocardioidaceae | Friedmanniella spumicola |
| LC026833 | Actinobacteria | Nocardioidaceae | Nocardioides ganghwensis |
| LC026826 | Actinobacteria | Nostocoida limicola | |
| LC026859 | Actinobacteria | Nostocoida limicola | |
| LC026900 | Actinobacteria | Nostocoida limicola | |
| LC026893 | Actinobacteria | Micromonosporaceae | Phytohabitans rumicis |
| LC026869 | Actinobacteria | Pseudonocardiaceae | Pseudonocardia alaniniphila |
| LC026878 | Actinobacteria | Pseudonocardiaceae | Pseudonocardia alaniniphila |
| LC026892 | Actinobacteria | Rubrobacteraceae | Rubrobacter xylanophilus |
| LC026853 | Armatimonadetes | Armatimonadaceae | Armatimonas rosea |
| LC026917 | Armatimonadetes | Armatimonadaceae | Armatimonas rosea |
| LC026852 | Bacteroidetes | Cytophagaceae | Adhaeribacter sp. TSX13-1 |
| LC026911 | Bacteroidetes | Bacteroidetes bacterium RG1-1 | |
| LC026922 | Bacteroidetes | Chitinophagaceae | Chitinophaga niabensis |
| LC026901 | Bacteroidetes | Cytophagaceae | Cytophagaceae bacterium MCCP1 |
| LC026823 | Bacteroidetes | Flammeovirgaceae | Flammeovirgaceae bacterium 311 |
| LC026851 | Bacteroidetes | Chitinophagaceae | Flavisolibacter ginsengisoli |
| LC026822 | Bacteroidetes | Chitinophagaceae | Flavisolibacter ginsengiterrae |
| LC026916 | Bacteroidetes | Chitinophagaceae | Flavisolibacter sp. MDT2-37 |
| LC026819 | Bacteroidetes | Cytophagaceae | Hymenobacter fastidiosus |
| LC026915 | Bacteroidetes | Cytophagaceae | Hymenobacter fastidiosus |
| LC026904 | Bacteroidetes | Cytophagaceae | Hymenobacter qilianensis |
| LC026940 | Bacteroidetes | Cytophagaceae | Hymenobacter sp. DG31A |
| LC026936 | Bacteroidetes | Cytophagaceae | Hymenobacter sp. R-37565 |
| LC026941 | Bacteroidetes | Cytophagaceae | Hymenobacter sp. R-37565 |
| LC026908 | Bacteroidetes | Cytophagaceae | Hymenobacter sp. VUG-A141a |
| LC026926 | Bacteroidetes | Cytophagaceae | Hymenobacter sp. VUG-A60a |
| LC026860 | Bacteroidetes | Cytophagaceae | Pontibacter diazotrophicus |
| LC026930 | Bacteroidetes | Cytophagaceae | Pontibacter korlensis |
| LC026842 | Bacteroidetes | Cytophagaceae | Pontibacter sp. MDT1-10-3 |
| LC026867 | Bacteroidetes | Cytophagaceae | Rhodocytophaga aerolata |
| LC026875 | Bacteroidetes | Cytophagaceae | Rhodocytophaga aerolata |
| LC026863 | Bacteroidetes | Cytophagaceae | Rufibacter tibetensis |
| LC026817 | Bacteroidetes | Chitinophagaceae | Segetibacter aerophilus |
| LC026820 | Bacteroidetes | Chitinophagaceae | Segetibacter aerophilus |
| LC026831 | Bacteroidetes | Chitinophagaceae | Segetibacter aerophilus |
| LC026836 | Bacteroidetes | Chitinophagaceae | Segetibacter aerophilus |
| LC026839 | Bacteroidetes | Chitinophagaceae | Segetibacter aerophilus |
| LC026848 | Bacteroidetes | Chitinophagaceae | Segetibacter aerophilus |
| LC026870 | Bacteroidetes | Chitinophagaceae | Segetibacter aerophilus |
| LC026879 | Bacteroidetes | Chitinophagaceae | Segetibacter aerophilus |
| LC026887 | Bacteroidetes | Chitinophagaceae | Segetibacter aerophilus |
| LC026906 | Bacteroidetes | Chitinophagaceae | Segetibacter aerophilus |
| LC026868 | Bacteroidetes | Cytophagaceae | Sporocytophaga myxococcoides |
| LC026876 | Bacteroidetes | Cytophagaceae | Sporocytophaga myxococcoides |
| LC026938 | Chloroflexi | Green non-sulfur bacterium AK-6 | |
| LC026942 | Chloroflexi | Oscillochloridaceae | Oscillochloris trichoides DG-6 |
| LC026902 | Cyanobacteria | Gloeocapsopsis crepidinum LEGE 06123 | |
| LC026835 | Cyanobacteria | Trichocoleus sociatus SAG 26.92 | |
| LC026847 | Deinococcus-Thermus | Deinococcaceae | Deinococcus deserti VCD115 |
| LC026872 | Deinococcus-Thermus | Deinococcaceae | Deinococcus sp. 4B6 |
| LC026881 | Deinococcus-Thermus | Deinococcaceae | Deinococcus sp. 4B6 |
| LC026837 | Deinococcus-Thermus | Trueperaceae | Truepera radiovictrix DSM 17093 |
| LC026889 | Deinococcus-Thermus | Trueperaceae | Truepera radiovictrix DSM 17093 |
| LC026913 | Deinococcus-Thermus | Trueperaceae | Truepera radiovictrix DSM 17093 |
| LC026914 | Deinococcus-Thermus | Trueperaceae | Truepera radiovictrix DSM 17093 |
| LC026811 | Firmicutes | Bacillaceae | Bacillus sp. IDA3504 |
| LC026855 | Firmicutes | Bacillaceae | Bacillus sp. IHB B 2252 |
| LC026827 | Firmicutes | Bacillaceae | Bacillus sp. ISO_06_Kulunda |
| LC026894 | Firmicutes | Bacillaceae | Bacillus sp. LCP70 |
| LC026886 | Firmicutes | Carnobacteriaceae | Carnobacterium sp. ARCTIC-P35 |
| LC026849 | Firmicutes | Paenibacillaceae | Paenibacillus sp. 6M01 |
| LC026937 | Firmicutes | Planococcaceae | Planococcus antarcticus |
| LC026939 | Firmicutes | Planococcaceae | Planococcus maritimus |
| LC026912 | Firmicutes | Planococcaceae | Planococcus sp. B17 |
| LC026829 | Firmicutes | Planococcaceae | Planococcus sp. PAMC 21323 |
| LC026832 | Firmicutes | Planococcaceae | Planococcus sp. PAMC 21323 |
| LC026821 | Firmicutes | Planococcaceae | Planococcus sp. YIM C738 |
| LC026910 | Firmicutes | Planococcaceae | Planococcus sp. YIM C738 |
| LC026920 | Firmicutes | Planococcaceae | Planomicrobium glaciei |
| LC026828 | Firmicutes | Planococcaceae | Planomicrobium koreense |
| LC026888 | Firmicutes | Planococcaceae | Planomicrobium sp. XN13 |
| LC026890 | Firmicutes | Planococcaceae | Planomicrobium sp. XN13 |
| LC026812 | Firmicutes | Veillonellaceae | Sporomusa paucivorans |
| LC026884 | Firmicutes | Thermoactinomycetaceae | Thermoactinomyces sp. T36 |
| LC026885 | Nitrospirae | Nitrospiraceae | Nitrospira sp. |
| LC026943 | Planctomycetes | Planctomycetaceae | Planctomycetaceae bacterium LX124 |
| LC026935 | Planctomycetes | Planctomycetaceae | Planctomycetaceae bacterium WSF3-27 |
| LC026830 | Proteobacteria | Rhodobacteraceae | Agaricicola taiwanensis |
| LC026862 | Proteobacteria | Alteromonadaceae | Alteromonas sp. Gp-4-13.1 |
| LC026898 | Actinobacteria | Micrococcaceae | Arthrobacter phenanthrenivorans |
| LC026834 | Proteobacteria | Rhodospirillaceae | Azospirillum sp. LH-CAB12 |
| LC026850 | Proteobacteria | Rhodospirillaceae | Azospirillum sp. LH-CAB12 |
| LC026905 | Proteobacteria | Bacterium Ellin5074 | |
| LC026816 | Proteobacteria | Candidatus Entotheonella palauensis | |
| LC026874 | Proteobacteria | Acetobacteraceae | Craurococcus roseus |
| LC026883 | Proteobacteria | Acetobacteraceae | Craurococcus roseus |
| LC026928 | Proteobacteria | Delta proteobacterium LX33 | |
| LC026825 | Proteobacteria | Oxalobacteraceae | Massilia sp. B48 |
| LC026896 | Proteobacteria | Phyllobacteriaceae | Nitratireductor sp. ZZ-1 |
| LC026919 | Proteobacteria | Nitrosomonadaceae | Nitrosomonas sp. Is79A3 |
| LC026909 | Proteobacteria | Oxalobacteraceae | Oxalobacter sp. W1.09-142 |
| LC026927 | Proteobacteria | Polyangiaceae | Polyangium fumosum |
| LC026814 | Proteobacteria | Rhodobacteraceae | Rubellimicrobium mesophilum DSM 19309 |
| LC026861 | Proteobacteria | Rhodobacteraceae | Rubellimicrobium roseum |
| LC026813 | Proteobacteria | Rhodospirillaceae | Skermanella sp. Py-2-1 |
| LC026895 | Proteobacteria | Rhodospirillaceae | Skermanella sp. Py-2-1 |
| LC026923 | Proteobacteria | Rhodospirillaceae | Skermanella sp. Py-2-1 |
| LC026929 | Proteobacteria | Sorangiineae | Sorangiineae bacterium SBSr006 |
| LC026873 | Proteobacteria | Sphingomonadaceae | Sphingomonas sediminicola |
| LC026882 | Proteobacteria | Sphingomonadaceae | Sphingomonas sediminicola |
| LC026865 | Unclassified | Bacterium Ellin6505 | |
| LC026934 | Unclassified | Bacterium LWQ8 | |
| LC026918 | Unclassified | Bacterium LY17 | |
| LC026846 | Unclassified | Bacterium WHC1-2 | |
| LC026854 | Unclassified | Bacterium YC-LK-LKJ4 | |
| LC026858 | Unclassified | Unidentified eubacterium clone BSV87 | |
| LC026824 | Verrucomicrobia | Verrucomicrobia bacterium WY51 |
In addition, we obtained several clones which were detected in the both DGGE analysis and clone library but not in the T-RFLP analysis; three clones of Deinococcus sp. (atmospheric bacteria; one was D. deserti and the others were unclassified to species level), as well as clones of soil bacteria such as three clones of Skermanella sp. (all unclassified to species level), seven clones of Planococcus sp. (one was P. antarcticus, one was P. maritimus and five were unclassified to species level) and eight clones of Hymenobacter sp. (two clones were H. fastidiosus, one clone was H. qilianensis and five clones were unclassified to species level). Also, as shown in Table 4, Segetibacter aerophilus (Bacteroidetes), which was first isolated from Asian dust samples collected in Korea with R2A agar,46) was dominant and occupied 7.5% of the clone library.
As previously mentioned, the source region of Asian dust particles during this Asian dust season was determined to be the Gobi Desert by HYSPLIT back trajectory analysis. We have analyzed the bacterial community structure of desert soil of the Gobi Desert26) and found it to be composed of Proteobacteria (36%), Actinobacteria (24%), Bacteroidetes (19%) and Firmicutes (6%). The bacterial community structure of Asian dust particles collected in Beijing was rather different from that of the dust source, the Gobi Desert. We have also analyzed bacterial community composition of Asian dust samples collected in Japan (Tottori Prefecture), 3000–5000 km from the dust source region26) in spring. These Asian dust particles were collected at 900 m altitude by a sampler set in an airplane. The major phyla identified in this Asian dust sample were Firmicutes (30%), Bacteroidetes (24%) and Actinobacteria (23%), and Proteobacteria (12%). These data demonstrate that the ratio of Firmicutes gradually increased from dust source region (6%) to Beijing (14%) and Tottori (30%), while Proteobacteria gradually decreased from dust source region (36%) to Beijing (18%) and to Tottori (12%).
Smith et al. analyzed the community structure of airborne bacteria transported to North America (Mt. Bachelor Observatory, Oregon; altitude of 2800 m) by Asian dust events,47) and they reported that the dominant bacterial phyla in their aerosol samples were Firmicutes and Actinobacteria. The distances are approximately 1500 km from Beijing to Tottori and 7500 km from Beijing to Mt. Bachelor Observatory. Most bacteria transported by aeolian dust will be stressed by the conditions encountered during atmospheric transport (UV exposure, reduced nutrient availability and desiccation). Firmicutes are known for desiccation-tolerant and thus the percentages of this phylum was probably increased during long-range transportation of Asian dust particles.
In this study, we visualized and quantified bacterial cells on Asian dust particles collected in Beijing where heavy Asian dust particles fall at approximately 180 t/km2/year and the influence of these dusts on health and ecosystems is of particular concern. Bacterial numbers on Asian dust particles seemed to increase compared with those in dust source regions. Inter-seasonal variability of bacterial community structure in Asian dust samples was low and it was assumed that bacterial community structure on dust particles changed during transportation. Results obtained in this study indicated that Asian dust contains high numbers of bacterial cells and their potential influence on health and ecosystems was demonstrated because some of those detected bacteria, including intestinal bacteria, remained viable.
We collected Asian dust samples at the top of a building in Beijing and therefore local soil particles might be mixed in the collected samples. When one has to analyze only the transported aeolian dust particles, one should collect dust particles at high altitude using balloons, helicopters or airplanes to avoid contamination of soil particles from the ground.26) On the other hand, one can collect dust particles near the ground when the purpose of the study is evaluation of the influence of those particles on our health and our surrounding living environment, because not only the transported particles but soil particles lifted up by wind near the sampling site can have the influence. Microbes in aeolian dust should have a greater influence in downwind areas near the dust source. Continuous temporal and spatial analyses from dust source regions to downwind regions will help to estimate the impact of atmospherically transported bacteria on health and ecosystems in downwind areas.
This study was supported by the JSPS KAKENHI (25281030) and the Environment Research and Technology Development Fund of the Ministry of the Environment, Japan (B-0902).
The authors declare no conflict of interest.