Abstract
Herpes simplex virus type 1 (HSV-1) is a causative agent for a variety of diseases. Although antiherpetic drugs such as acyclovir have been developed to inhibit virus replication through interaction with DNA kinases, their continuous administration leads to an increase in the frequency of drug-resistant HSV-1, which is an important clinical issue that requires urgent solution. Recently, we reported that the sialylated O-linked sugar T antigen (sTn) and its attached peptide region (O-glycosylated sTn peptide) derived from the HSV-1 glycoprotein B (gB) protein inhibited HSV-1 infection by specifically targeting paired immunoglobulin-like type 2 receptor alpha (PILRα) in vitro. In this study, to further identify novel inhibitors of gB-mediated HSV-1 infection in vitro, we established a cell-based fusion assay for rapid drug screening. Chinese hamster ovary (CHO) cells were transfected with expression plasmids for HSV-1 gB, gD, gH, and gL, and T7 RNA polymerase, and were designated as the effector cells. The CHO-K1 cells stably expressing PILRα were transfected with the expression plasmid for firefly luciferase under the T7 promoter, and were designated as the target cells. The effector and target cells were co-cultured, and luminescence was measured when both cells were successfully fused. Importantly, we found that cell-to-cell fusion was specifically inhibited by O-glycosylated sTn peptide in a dose dependent manner. Our results suggested that this virus-free cell-based fusion assay system could be a useful and promising approach to identify novel inhibitors of gB-mediated HSV-1 infection, and will aid in the development of antiviral therapeutic strategies for HSV-1-associated diseases.
Herpesviridae is a family of DNA viruses that consists of nine human viruses belonging to the alphaherpesviridae (herpes simplex virus (HSV)-1, HSV-2, and varicella-zoster virus), betaherpesviridae (cytomegalovirus (CMV)), human herpes virus ((HHV)-6A, HHV-6B, and HHV-7), and gammaherpesviridae (Epstein–Barr virus (EBV) and HHV-8) subfamilies.1) HSV-1 is the causative agent of encephalitis, meningitis, myelitis, radiculitis, and keratitis.2) In addition, neonatal HSV-1 infection can also occur in utero, intrapartum, or postnatally.3) Once HSV-1 infects and enters the target cells, it establishes a latent infection, which results in a long-term asymptomatic state. However, viral reactivation may occur as a result of environmental and non-environmental stimuli such as an immunosuppressant state, stress, and ultraviolet light.4) Thus, HSV-1 infection remains a major global concern.
Antiherpetic drugs such as acyclovir (ACV) or penciclovir (PCV), and their structure- or function-related derivatives have been produced as first-line drugs.5) ACV, phosphorylated by viral thymidine kinase and cellular kinases, acts as a substrate for viral DNA polymerase, and is incorporated into the viral DNA leading to the termination of viral DNA synthesis.6) Although these inhibitors exhibit anti-viral effects on virus replication, continuous administration of them as long-term prophylaxis and treatment frequently results in the generation of drug-resistant mutant HSV-1, which is an important clinical issue needing to be urgently resolved, especially among immunocompromised patients.7) Viral resistance to ACV could be caused by mutations in the thymidine kinase gene.7) Recently, FIT-039, an inhibitor of cyclin-dependent kinase (CDK) 9 was developed to inhibit HSV-1 and -2, as well as other DNA viruses such as CMV and adenovirus.8) Furthermore, FIT-039 exhibited inhibitory effects to ACV-resistant HSV-1 infection.8) Thus, targeting of not only viral genes or products but also cellular factors should be considered.
Based on this idea, blocking the attachment and entry steps of viral infection into human target cells is also an attractive therapeutic strategy; an advantage of this concept is that it could prevent spread of the virus without killing virus-infected cells, like ACV, which could be beneficial for tissues lacking regenerative capacity (e.g. nervous system tissues). Similar to virus entry into the target cells, an interaction between the infected and uninfected cells induces cell fusion, which also promotes the spread of HSV-1 between cells. Such cell fusions are observed for a variety of viruses.9) In fact, it has been reported that the sulfated oligosaccharide mixtures, muparfostat (previously known as PI-88) and cyanovirin-N, can impair viral glycoprotein-induced cell fusion.10,11) In addition, n-docosanol (Abreva) is another fusion inhibitor that has been approved by the Food and Drug Administration (FDA) and marketed for the treatment of recurrent herpes simplex labialis.12,13) In recent years, it has also been shown that antimicrobial peptides (AMP),14) or other synthetic peptides that bind to 3-OS HS exhibited anti-HSV-1 effects during HSV-1 infection in vivo.15,16) The use of peptides has several advantages compared with chemical compounds; they can exert a broad spectrum of activity on different microorganisms, and they are highly specific and effective. However, their ability to cross the cell membrane is limited, therefore the use of peptides may be suitable to act on cell membranes. In these cases, they can be used to specifically inhibit glycoprotein-receptor interactions; among the 12 glycoproteins encoded in HSV-1, four glycoproteins, gB, gD, gH, and gL, are cooperatively utilized for attachment and cell entry.17) Specifically, gB plays a critical role for entry into the target cells.18,19) Entry consists of a multistep process involving several cellular receptors, paired immunoglobulin-like type 2 receptor alpha (PILRα),20) as well as non-muscle myosin IIA (NM-IIA)21) and myelin associated glycoprotein (MAG),22) which have been shown to interact with gB. Since PILRα is an inhibitory receptor, and its binding to gB could induce immune suppression followed by acceleration of immune evasion by HSV-1, inhibition of the gB-PILRα interaction could be a promising target to reduce the spread of viruses in vivo. The inhibitory effects of peptides (corresponding to amino acid residues 491–514) derived from gB during HSV-1 infection have been demonstrated when investigating the role of the critical fusogenic domain of gB in the entry process.23) Akkarawongsa et al. also reported the inhibition of HSV-1 entry by gB-derived peptides.24) Recently, we identified the sialylated O-linked sugar T antigen (sTn) and its attached peptide region, derived from gB (O-glycosylated sTn peptide; corresponding to amino acid residues 50–56), that effectively inhibited HSV-1 infection by binding to PILRα in vitro,25) thus, this O-glycosylated sTn peptide could be a promising lead molecule for the inhibition of gB-mediated HSV-1 infection.
In this study, to further identify and evaluate novel antivirals for gB-mediated HSV-1 infection in vitro, we established a virus-free cell-based fusion assay as a rapid and simple drug screening method rather than the virus infection-induced plaque assay. In the cell fusion assay presented here, we utilized the O-glycosylated sTn peptide to specifically block the interaction of gB with PILRα. Here we show that this cell-based fusion assay can be a rapid and reliable method for identifying novel antivirals, such as small chemical compounds, synthetic peptides, and monoclonal antibodies, of gB-mediated HSV-1 entry.
MATERIALS AND METHODS
CellsChinese hamster ovary (CHO) cells were grown in Ham’s F-12 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), penicillin (100 units/mL), and streptomycin (100 µg/mL). G418 (250 µg/mL) (Wako Pure Chemical Industries, Ltd., Osaka, Japan) was added to the culture of CHO-K1 cells stably expressing human PILRα (CHO-hPILRα).
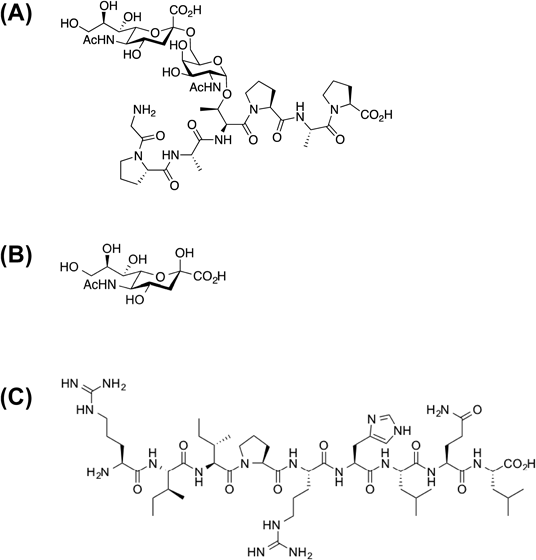
Chemical Synthesis of O-Glycosylated Peptides and Control MaterialsThe peptide, whose sequence is arginine–isoleucine–isoleucine–proline–arginine–histidine–leucine–glutamine–leucine (RIIPRHLQL), designated as the control peptide, and O-glycosylated sTn derivative were synthesized by unpublished methods (manuscript in preparation) (Fig. 1). They were dissolved in water at a concentration of 10 mM.
In Vitro Cell Fusion AssayCHO and CHO-hPILRα cells were seeded at a density of 5×105 cells in 6-well plates, and 1×104 cells in 96-well plates, and were incubated for 12 h at 37°C. The expression plasmids for T7 RNA polymerase (pCAG T7)26) and either HSV-1 (KOS strain) glycoproteins gB, gD, gH, and gL (pPEP98-gB, pPEP99-gD, pPEP100-gH, and pPEP101-gL, respectively),27) or HSV-1 (F strain) glycoproteins20) were transfected into CHO cells, and were designated as the effector cells. The expression plasmid for the firefly luciferase gene under the control of the T7 promoter (pT7EMCLuc)26) was transfected into CHO-hPILRα cells, and were designated as the target cells. Both types of transfection was carried out using FuGENE 6 Transfection Reagent (Roche Applied Science, Pleasanton, CA, U.S.A.), and were incubated for 12 h at 37°C. The target cells were pre-treated with 50–200 µM of the O-glycosylated sTn peptide, sialic acid, or control peptide (RIIPRHLQL) for 2 h at 37°C. The effector cells were detached with 0.25% trypsin-ethylenediaminetetraacetic acid (EDTA), and 2×104 of the effector cells were overlaid to the target cells, and further incubated for 12–18 h at 37°C. The cells were lysed with lysis buffer (25 mM Tris–Cl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 5% glycerol) supplemented with Pierce Protease Inhibitor Mini Tablets (Thermo Fisher Scientific, Waltham, MA, U.S.A.) for 20 min, and Steady-Glo Luciferase Assay Substrate (Promega, Madison, WI, U.S.A.) was added to the lysates. The absorbance of the lysate samples was measured using an EnSpire microplate reader (PerkinElmer, Inc., Waltham, MA, U.S.A.).
In Vitro Cell Proliferation AssayCHO-hPILRα cells were seeded at a density of 5×103 cells in the presence of 50–200 µM of O-glycosylated sTn peptide, sialic acid, or control peptide, and incubated for 24 h at 37°C. The cells were further incubated with Cell Proliferation Reagent WST-1 (Roche Diagnostics, Deutschland, GmbH) for 2 h at 37°C. The absorbance of the samples was measured at 450 and 550 nm (reference wave length) using a microplate reader.
Flow Cytometric AnalysisThe expression plasmids for T7 RNA polymerase and HSV-1 glycoproteins gB, gD, gH, and gL were transfected into CHO cells as described above. Twenty four hours after transfection, the CHO cells were stained with mouse monoclonal antibodies to gB (clone H1817; Virusys), gD (clone DL6; Santa Cruz Biotechnology), and gH (clone 52-S; American Type Culture Collection), and isotype immunoglobulin Gs (IgGs) (Millipore) as negative controls. The expression of HSV-1 glycoproteins was examined by using a FACS Calibur flow cytometer (BD Biosciences). Background-corrected mean fluorescence intensity was determined.
RESULTS AND DISCUSSION
We have previously confirmed that gB-derived O-glycosylated sTn peptide interacts with PILRα through surface plasmon resonance (SPR) competition assays.25) To evaluate the non-specific effects of O-glycosylated sTn peptide on cell fusion, we chemically synthesized a control peptide RIIPRHLQL (Fig. 1). We did not observe any interaction of the controls with PILRα when analyzed by SPR (data not shown). Based on interaction assays at the molecular level, these results strongly suggested that the O-glycosylated sTn peptide may be an attractive candidate as an HSV-1 inhibitor for interfering with the gB-PILRα interaction.
To establish an in vitro system for evaluating the inhibitory effects of antivirals on gB-mediated HSV-1 infection, we developed a cell-to-cell fusion assay (as illustrated in Fig. 2). The target CHO-hPILRα cells were prepared by transient transfection with a plasmid encoding firefly luciferase, while the effector cells were transiently transfected with a plasmid encoding HSV-1 (KOS strain) glycoproteins and another plasmid that expresses T7 RNA polymerase. Similar assays have been used to investigate cell fusion mechanisms in other herpes viruses, such as HHV-8 and EBV,28,29) suggesting that the fusion assay should be a reliable procedure for screening herpes virus inhibitors. However, the advantage of our assay compared to those used in previous studies is that we used CHO cells stably expressing PILRα as target cells instead of cells expressing several distinct HSV-1 receptors. This enabled us to evaluate gB-specific inhibitors by eliminating the potential interaction of gB with other receptors. Moreover, after co-cultivation of the target and effector cells for 12 h, the luminescence of cells treated with water was 1.037×105 ±4.268×103 relative luciferase units (RLU). In contrast, background luminescence, as determined by co-culturing nontransfected target cells and CHO cells, was less than 1.0×103 RLU (Fig. S1). These results were reproducible, and the high detection sensitivity would be very useful for drug screening.
The plasmids encoding HSV-1 (F strain) glycoproteins were also used for the fusion assay, but the luminescence of cells transfected with HSV-1 (F strain) glycoproteins was lower than those transfected with HSV-1 (KOS strain) glycoproteins (Fig. S2). This indicated that cell fusion via HSV-1 (F strain) glycoprotein-PILRα interactions may not be efficient. Flow cytometric analysis demonstrated that expression of glycoproteins on CHO cells transfected with HSV-1 (F strain) was lower than that of the HSV-1 (KOS strain) (Fig. S3). Thus, the level of HSV-1 (KOS strain) glycoprotein expression achieved using this transient transfection method makes this assay sensitive enough to screen for HSV-1 inhibitors.
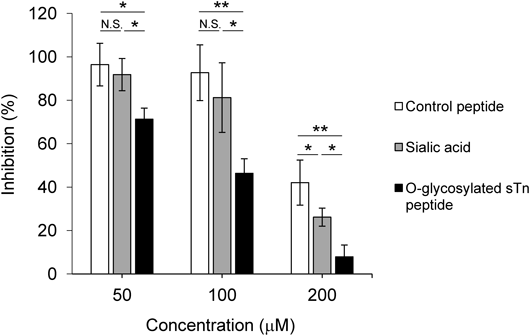
To investigate the effect of inhibitors on the fusion assay, the target CHO-hPILRα cells transiently transfected with the plasmid encoding firefly luciferase were pretreated with 50–200 µM of the O-glycosylated sTn peptide, sialic acid, or a control peptide. The target cells were then co-cultured with the effector CHO cells transiently transfected with the plasmid encoding HSV-1 (KOS strain) glycoproteins and the plasmid encoding T7 RNA polymerase. None of the three agents tested caused cytotoxicity in CHO-hPILRα cells when assessed by cell proliferation assays, even at 200 µM (Fig. 3). Notably, we found that the luminescence of cells treated with the O-glycosylated sTn peptide significantly decreased in a dose-dependent manner (inhibition of cell fusion was ca. 29% at 50 µM, ca. 54% at 100 µM, and ca. 92% at 200 µM compared to cells treated with only vehicle). This suggested that the O-glycosylated sTn peptide specifically blocks cell-to-cell fusion (Fig. 4).
The concentration of O-glycosylated sTn peptide in this study was higher than previously reported.25) It is likely that the concentration of inhibitors in a cell-based assay is higher than that in a cell-free infection assay.10) It is of note that the O-glycosylated sTn peptide could exhibit inhibitory effects on both cell-free virus infection and cell–cell virus transmission in vivo. The luminescence of cells treated with sialic acid or control peptides at 200 µM was significantly decreased, indicating nonspecific binding of these materials to the cell membrane upon cell fusion (Fig. 4). Therefore, using appropriate control materials that do not cause cytotoxicity in the target cells (as shown in Fig. 3) is necessary to distinguish specific versus nonspecific binding. Collectively, these results strongly suggested that our virus-free cell-based fusion assay is a reliable, simple, and rapid procedure to evaluate the effects of antiviral inhibitors on gB-mediated HSV-1 infection in vitro.
Identification of novel antiviral compounds should be dependent on an in vitro primary assay for high-throughput screening (HTS) of the compound libraries.30) In addition, structure-based drug design (SBDD) could be a powerful tool,31) especially for identifying novel inhibitors of viruses.32) We are currently performing this cell-based fusion assay for the screening of small chemical compounds from the library against HSV-1 infection, in combination with SBDD. The chemical compounds in the library are dissolved with dimethyl sulfoxide (DMSO), and we have confirmed that the final concentration of DMSO to a maximum of 1% in culture is tolerable for the cell fusion assay, as evaluated by a decrease of luminescence (data not shown). In conclusion, as an in vitro primary HTS, our cell-based fusion assay may be a useful tool for identifying promising lead compounds with similar effects as the O-glycosylated sTn peptide to block the gB-mediated HSV-1 entry.
Acknowledgments
We thank Dr. Yoshiharu Matsuura for providing the expression plasmids, pCAGT7 and pT7EMCLuc, and Dr. Patricia Spear for the expression plasmids, pPEP98-gB, pPEP99-gD, pPEP100-gH, and pPEP101-gL. This work was partially supported by Platform for Drug Discovery, Informatics, and Structural Life Science by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, a Grant-in-Aid (A) for Scientific Research by the Japan Society for the Promotion of Science (JSPS), and Bioconjugate project of Hokkaido University (to K.M.).
Conflict of Interest
The authors declare no conflict of interest.
Supplementary Materials
The online version of this article contains supplementary materials.
Fig. S1. Luminescence in Cell Fusion Assay
In the cell fusion assay, the expression plasmids for T7 RNA polymerase and HSV-1 (KOS strain) glycoproteins gB, gD, gH, and gL were transfected into CHO cells (Plasmid (+)), and were designated as the effector cells. CHO cells without any transfection (Plasmid (−)) were used as the effector cells to check the background luminescence. Bars indicate mean values±S.E.M. Data are representative of three independent experiments.
Fig. S2. Comparison of Glycoproteins from Different HSV-1 Strains on Cell Fusion
Experimental procedure is described in Materials and Methods. Bars indicate mean values±S.E.M. Statistically significant differences are shown as p values (** p<0.01). Data are representative of three independent experiments.
Fig. S3. Expression of HSV-1 Glycoproteins on the Effector CHO Cells
The expression plasmids for T7 RNA polymerase and glycoproteins gB, gD, gH, and gL of HSV-1 KOS strain or F strain were transfected into CHO cells. The expression of gB, gD, and gH–gL complex on the CHO cells was evaluated using a FACS Calibur flow cytometer. Black open histograms indicate cells stained with monoclonal antibodies to the indicated HSV-1 glycoproteins. Gray filled histograms represent cells stained with isotype control IgGs.
REFERENCES
- 1) Piret J, Boivin G. Antiviral drug resistance in herpesviruses other than cytomegalovirus. Rev. Med. Virol., 24, 186–218 (2014).
- 2) Roizman B, Knipe DM, Whitley RJ. Herpes simplex viruses. Fields Virology (Knipe DM, Howley PM eds.) 5th Edition, Lippincott, Williams, & Wilkins, Philadelphia, P.A., pp. 2501–2602 (2007).
- 3) Whitley RJ, Roizman B. Herpes simplex virus infections. Lancet, 357, 1513–1518 (2001).
- 4) Roizman B, Whitley RJ. An inquiry into the molecular basis of HSV latency and reactivation. Annu. Rev. Microbiol., 67, 355–374 (2013).
- 5) Kukhanova MK, Korovina AN, Kochetkov SN. Human herpes simplex virus: life cycle and development of inhibitors. Biochemistry (Mosc), 79, 1635–1652 (2014).
- 6) Elion GB, Furman PA, Fyfe JA, de Miranda P, Beauchamp L, Schaeffer HJ. Selectivity of action of an antiherpetic agent, 9-(2-hydroxyethoxymethyl)guanine. Proc. Natl. Acad. Sci. U.S.A., 74, 5716–5720 (1977).
- 7) Piret J, Boivin G. Resistance of herpes simplex viruses to nucleoside analogues: mechanisms, prevalence, and management. Antimicrob. Agents Chemother., 55, 459–472 (2011).
- 8) Yamamoto M, Onogi H, Kii I, Yoshida S, Iida K, Sakai H, Abe M, Tsubota T, Ito N, Hosoya T, Hagiwara M. CDK9 inhibitor FIT-039 prevents replication of multiple DNA viruses. J. Clin. Invest., 124, 3479–3488 (2014).
- 9) Harrison SC. Viral membrane fusion. Virology, 479–480, 498–507 (2015).
- 10) Ekblad M, Adamiak B, Bergstrom T, Johnstone KD, Karoli T, Liu L, Ferro V, Trybala E. A highly lipophilic sulfated tetrasaccharide glycoside related to muparfostat (PI-88) exhibits virucidal activity against herpes simplex virus. Antiviral Res., 86, 196–203 (2010).
- 11) Tiwari V, Shukla SY, Shukla D. A sugar binding protein cyanovirin-N blocks herpes simplex virus type-1 entry and cell fusion. Antiviral Res., 84, 67–75 (2009).
- 12) Pope LE, Marcelletti JF, Katz LR, Lin JY, Katz DH, Parish ML, Spear PG. The anti-herpes simplex virus activity of n-docosanol includes inhibition of the viral entry process. Antiviral Res., 40, 85–94 (1998).
- 13) Marcelletti JF. Synergistic inhibition of herpesvirus replication by docosanol and antiviral nucleoside analogs. Antiviral Res., 56, 153–166 (2002).
- 14) Galdiero S, Falanga A, Tarallo R, Russo L, Galdiero E, Cantisani M, Morelli G, Galdiero M. Peptide inhibitors against herpes simplex virus infections. J. Pept. Sci., 19, 148–158 (2013).
- 15) Tiwari V, Liu J, Valyi-Nagy T, Shukla D. Anti-heparan sulfate peptides that block herpes simplex virus infection in vivo. J. Biol. Chem., 286, 25406–25415 (2011).
- 16) Tiwari V, Tarbutton MS, Shukla D. Diversity of heparan sulfate and HSV entry: basic understanding and treatment strategies. Molecules, 20, 2707–2727 (2015).
- 17) Antoine TE, Park PJ, Shukla D. Glycoprotein targeted therapeutics: a new era of anti-herpes simplex virus-1 therapeutics. Rev. Med. Virol., 23, 194–208 (2013).
- 18) Kawaguchi Y. Herpes simplex virus (HSV). Uirusu, 60, 187–196 (2010).
- 19) Satoh T, Arase H. HSV-1 infection through inhibitory receptor, PILRα. Uirusu, 58, 27–36 (2008).
- 20) Satoh T, Arii J, Suenaga T, Wang J, Kogure A, Uehori J, Arase N, Shiratori I, Tanaka S, Kawaguchi Y, Spear PG, Lanier LL, Arase H. PILRα is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell, 132, 935–944 (2008).
- 21) Arii J, Goto H, Suenaga T, Oyama M, Kozuka-Hata H, Imai T, Minowa A, Akashi H, Arase H, Kawaoka Y, Kawaguchi Y. Non-muscle myosin IIA is a functional entry receptor for herpes simplex virus-1. Nature, 467, 859–862 (2010).
- 22) Suenaga T, Satoh T, Somboonthum P, Kawaguchi Y, Mori Y, Arase H. Myelin-associated glycoprotein mediates membrane fusion and entry of neurotropic herpesviruses. Proc. Natl. Acad. Sci. U.S.A., 107, 866–871 (2010).
- 23) Galdiero S, Vitiello M, D’lsanto M, Falanga A, Cantisani M, Browne H, Pedone C, Galdiero M. The identification and characterization of fusogenic domains in herpes virus glycoprotein B molecules. ChemBioChem, 9, 758–767 (2008).
- 24) Akkarawongsa R, Pocaro NE, Case G, Kolb AW, Brandt CR. Multiple peptides homologous to herpes simplex virus type 1 glycoprotein B inhibit viral infection. Antimicrob. Agents Chemother., 53, 987–996 (2009).
- 25) Kuroki K, Wang J, Ose T, Yamaguchi M, Tabata S, Maita N, Nakamura S, Kajikawa M, Kogure A, Satoh T, Arase H, Maenaka K. Structural basis for simultaneous recognition of an O-glycan and its attached peptide of mucin family by immune receptor PILRα. Proc. Natl. Acad. Sci. U.S.A., 111, 8877–8882 (2014).
- 26) Okuma K, Nakamura M, Nakano S, Niho Y, Matsuura Y. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology, 254, 235–244 (1999).
- 27) Pertel PE, Fridberg A, Parish ML, Spear PG. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH–gL requires a gD receptor but not necessarily heparan sulfate. Virology, 279, 313–324 (2001).
- 28) Pertel PE. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J. Virol., 76, 4390–4400 (2002).
- 29) Haan KM, Lee SK, Longnecker R. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein–Barr virus-induced membrane fusion. Virology, 290, 106–114 (2001).
- 30) Kakisaka M, Sasaki Y, Yamada K, Kondoh Y, Hikono H, Osada H, Tomii K, Saito T, Aida Y. A novel antiviral target structure involved in the RNA binding, dimerization, and nuclear export functions of the influenza A virus nucleoprotein. PLoS Pathog., 11, e1005062 (2015).
- 31) Lounnas V, Ritschel T, Kelder J, McGuire R, Bywater RP, Foloppe N. Current progress in structure-based rational drug design marks a new mindset in drug discovery. Comput. Struct. Biotechnol. J., 5, e201302011 (2013).
- 32) Cancellieri M, Bassetto M, Widjaja I, van Kuppeveld F, de Haan CAM, Brancale A. In silico structure-based design and synthesis of novel anti-RSV compounds. Antiviral Res., 122, 46–50 (2015).