2016 年 39 巻 7 号 p. 1211-1215
2016 年 39 巻 7 号 p. 1211-1215
Nephronectin (Npnt), known to be a ligand of integrin α8β1, plays important roles in the development and function of various tissues, including those of the kidneys, liver, bones, and muscles. In previous studies, we showed that the expression of Npnt mRNA was regulated by various cytokines, including transforming growth factor-β (TGF-β), tumor necrosis factor-α (TNF-α), and oncostatin M (OSM), and that over-expression of Npnt enhanced osteoblast differentiation. In this study, we found that bone morphogenic protein-2 (BMP-2), known as an osteogenesis inducing cytokine, strongly up-regulated the expression of Npnt mRNA in a murine skeletal muscle cell line (C2C12) via the BMP-SMAD signaling pathway.
Nephronectin (Npnt), identified as a ligand of integrin α8β1, plays essential roles in the development and functions of various organs and tissues, including those of the kidneys, liver, bones, and muscles.1–3) Recently, we reported that the expression of Npnt mRNA was down-regulated by various cytokines, including transforming growth factor-β (TGF-β), tumor necrosis factor-α (TNF-α), and oncostatin M (OSM).4–6) Moreover, in regard to studies of osteoblast extracellular matrix protein, some reports have revealed that over-expression of Npnt promoted osteoblast differentiation.5–7)
C2C12 cells are derived from satellite cells obtained from murine skeletal muscle tissues and widely used for in vitro differentiation experiments, as they can be induced to either multinucleated myotubes after withdrawal of serum from the culture medium or a differentiation pathway that leads to osteoblastic lineage cells by introduction of osteogenic bone morphogenetic proteins (BMPs).8)
BMPs are cytokines that belong to the transforming growth factor-β (TGF-β) superfamily. Various types of cells, including bone marrow stromal cells, chondrocytes, and osteoblasts, generate BMPs and are subsequently affected by them for differentiation, proliferation, and other functions.9,10) A previous study found that BMPs implanted into bone defects initiated bone healing and increased the stiffness of regenerating bone tissues in vivo.11) BMPs bind to complexes composed of type I receptors, including activin receptor-like kinase 1 (ALK1), 2, 3, and 6, as well as type II receptors, to which ActRIIa/b and BMPRII belong. After binding with BMPs, the type I receptor becomes phosphorylated, then activates the canonical BMP-SMAD signaling pathway as well as non-SMAD signaling pathways including mitogen-activated protein kinase (MAPK), which induces osteoblast differentiation.12)
In the present study, we investigated the relationship between Npnt and BMPs in C2C12 cells, and found that BMP-2 significantly up-regulated the expression of Npnt mRNA. Our findings suggest a role of Npnt in osteoblast differentiation induced by BMP-2.
C2C12 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (high glucose) with L-glutamine and Phenol Red medium (Wako Pure Chemical Industries, Ltd., Osaka, Japan; Cat. No. 044-29765), supplemented with 15% fetal bovine serum (FBS) (Life Technologies, U.S.A.; Cat. No. 10437) and 1% penicillin-streptomycin at 37°C in a CO2 incubator (5% CO2, 95% air). For the experiments, cells were seeded into 6-well (Thermo Scientific Inc., U.S.A.; Cat. No. 140675) or 96-well (Thermo Scientific Inc., Cat. No. 167008) plates.
ReagentsRecombinant human bone morphogenetic protein 2 (rhBMP-2) (Cat. No. 355-BM), rhBMP-3 (Cat. No. 113-BP), rhBMP-4 (Cat. No. 314-BP), rhBMP-6 (Cat. No. 507-BP), and rhBMP-7 (Cat. No. 354-BP) were purchased from R&D Systems, Inc., U.S.A.; LDN193189 hydrochloride (Cat. No. 124-06011, Wako Pure Chemical Industries, Ltd.) was used as an inhibitor of the BMP type I receptors ALK 2, ALK3, and ALK6 (IC50 ≤ 5, 30, and 150 nM, respectively).13)
Quantitative Real-Time Polymerase Chain Reaction (PCR)Total cellular RNA was isolated using TRIzol reagent (Life Technologies; Cat. No. 15596018) or a SuperPrepTM Cell Lysis&RT Kit for qPCR (TOYOBO, Ltd., Japan; Code No. SCQ-101S), then each 2 µg of cellular RNA was reverse-transcribed using Super Script III (Life Technologies, Cat. No. 18080-044). Quantitative real-time PCR was performed with a StepOne™ Real Time PCR System (Applied Biosystems, U.S.A.) using SYBR Green Fast PCR Master Mix (Applied Biosystems) with the following specific PCR primers.
Glyceraldehyde-3-phosphate dehydrogenase (Gapdh): 5′-AAA TGG TGA AGG TCG GTG TG-3′,5′-TGA AGG GGT CGT TGA TGG-3′. Npnt: 5′-CAC GAG TAA TTA CGG TTG ACA ACA G-3′, 5′-CTG CCG TGG AAT GAA CAC AT-3′. Smad4: 5′-GCT TGG GTC AAC TCT CCA ATG-3′, 5′-TGT GCA ACC TCG CTC TCT CA-3′.
Western BlottingCells were lysed using 10 mm Tris–HCl (pH 7.8) containing 1% Nonidet P-40, 0.15 m NaCl, and a protease inhibitor mixture containing ethylenediaminetetraacetic acid (EDTA) (Roche Applied Science, U.S.A.). Lysates were cleared by centrifugation at 13000×g for 10 min, then placed onto 7.5% sodium dodecyl sulfate (SDS) polyacrylamide gels and electrophoresed following dilution with Sample Buffer Solution with Reducing Reagent (6x) for SDS-polyacrylamide gel electrophoresis (PAGE) (Nacalai Tesque, Kyoto, Japan; Cat. No. 09499-14). After transferring, polyvinylidene difluoride (PVDF) membranes were blocked with 0.1% Tween20-TBS containing 1% skim milk for 1 h and incubated with the primary antibody overnight at 4°C. Antibodies against the following proteins were used for Western blotting analysis: Smad1 (1/1000, Cell Signaling Technology, U.S.A.; #9743), Smad5 (1/625, Cell Signaling Technology; #12534), phospho-Smad1/5 (1/1000, Cell Signaling Technology; #9516), Smad4 (1/1000, Cell Signaling Technology; #9515), Npnt (1/500, R&D Systems, Inc. #AF4298), and actin (1/2000, SIGMA Life Science, U.S.A.; #A5060). Anti-rabbit immunoglobulin G (IgG), horseradish peroxidase linked whole antibody (1/2500, GE Healthcare, U.S.A.; Cat. No. NA934V), and peroxidase-conjugated anti-goat IgG (1/5000, Jackson ImmunoResearch, U.S.A.; #705-035-147) were used as secondary antibodies. All antibodies were placed in TBS-0.1% Tween20 containing 1% skim milk. To visualize antigenic bands, peroxidase reactions were developed using ECL Prime Western blotting Detection Reagent (GE Healthcare, U.S.A.; RPN2232).
Small Interfering RNA (siRNA) Knockdown of Gene ExpressionFor siRNA transfection, Lipofectamine RNAiMAX Reagent (Invitrogen, U.S.A.; #13778-150) was used. Both of Smad4 siRNA (Cell Signaling Technology; #12791) and StealthTM RNAi Negative Control Medium GC Duplex #2 (Invitrogen; PN12935112) were diluted by adding Opti-MEM (Gibco, U.S.A.; REF 31985-070) in different tubes. After Lipofectamine RNAiMAX Reagent was added, they were incubated at room temperature for 20 min. Next, each of the siRNA complex solutions was added into a medium containing 5×104 cells/mL, and the mixed solutions were placed into plates. siRNA was transfected at a final concentration of 60 nM. After a 48 h treatment, the cells were washed to remove the siRNA-containing medium and incubated in the culture medium containing 2.5% FBS in the presence or absence of 300 ng/mL of BMP-2 for an additional 48 h. StealthTM RNAi Negative Control Medium GC Duplex #2 (Invitrogen; PN12935112) was used for a negative control siRNA.
Luciferase Assay AnalysisId1 is used as a quick responsive gene for BMP-2 treatment and its response is mediated by binding of Smad4 to the BMP binding site in the Id1 gene. To examine the effect of smad4 siRNA, we transfected an Id1.0-luc reporter plasmid constructed in a previous report and a TK vector into C2C12 cells after 48 h of treatment with smad4 siRNA.14) Luminosity was detected using a Dual-Luciferase Reporter Assay System (Promega, U.S.A.; E1910) after 24 h of incubation with or without 300 ng/mL of BMP-2. The relative firefly luciferase reaction was normalized to the luminescent reactions of Renilla luciferase from the TK vector. A pGL4 [luc2/Neo] vector was used as the negative control vector.
Myosin Heavy Chain (MHC) and Alkaline Phosphatase (ALP) StainingTo examine C2C12 cell differentiation, we checked expression of MHC and activity of ALP. First, to analyze the expression of MHC, cells were fixed with 10% formaldehyde for 10 min at room temperature, then treated with an acetone–ethanol mixture (50 : 50, v/v) for 1 min and incubated with a mouse anti-MHC monoclonal antibody (MF-20; Developmental Studies Hybridoma Bank, Iowa, IA, U.S.A.) at room temperature for 1 h. After washing with PBS, cells were incubated for 30 min with a secondary antibody (#424134; Nichirei, Tokyo, Japan). To determine ALP activity, after washing with PBS, cells were exposed to an ALP activity solution containing 0.1 mg/mL of naphthol AS-MX phosphate (Sigma), 0.5% N,N-dimethylformamide (Wako Pure Chemical Industries, Ltd.), 2 mM MgCl2, and 0.6 mg/mL of fast blue BB salt (Sigma) in 0.1 mM Tris–HCl (pH 8.5) for 30 min at room temperature. After washing with PBS, MHC was visualized using an AEC substrate kit (#415184, Nichirei).
Statistical AnalysisAll results are expressed as the mean±standard deviation (S.D.). Statistical analysis results shown in Figs. 1, 2A, and 3A, and Supplementary Fig. 2B were obtained using one-way ANOVA, while those show in Figs. 2B, 4A, and B were obtained using a two-tailed Student’s t-test. p Values <0.05 were considered to be statistically significant.
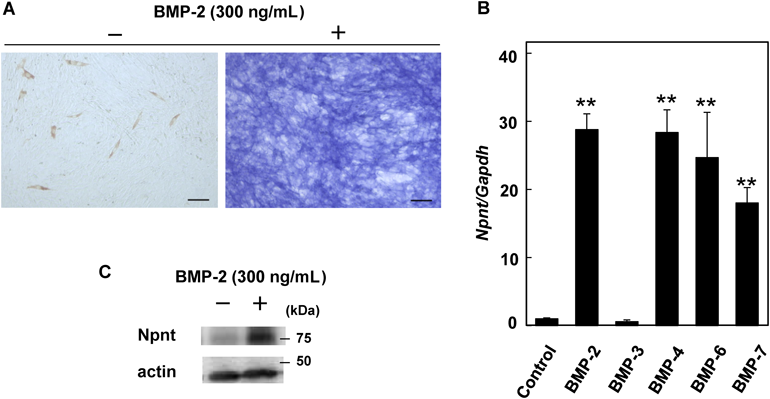
(A) Double staining of MHC and ALP in C2C12 cells cultured with or without BMP-2 for 48 h. MHC and ALP activity were used as markers for differentiation into myotubes and osteoblasts, respectively. All images are shown after staining and drying for 24 h. Scale bars represent 100 µm. (B) Up-regulation of Npnt mRNA expression by BMPs. Real-time PCR analysis was performed using cDNA from C2C12 cells treated for 48 h with 300 ng/mL of BMP-2, BMP-3, BMP-6, or BMP-7, or 240 ng/mL of BMP-4. Values are shown as the mean±S.D. of 3 samples folded over the mean of the group without BMP treatment. ** p<0.01, relative to level in cells without treatment (ANOVA). (C) Western blotting analysis of Npnt protein levels in cells treated with and without BMP-2. Cell lysates were collected after 48 h of incubation with or without 300 ng/mL of BMP-2.
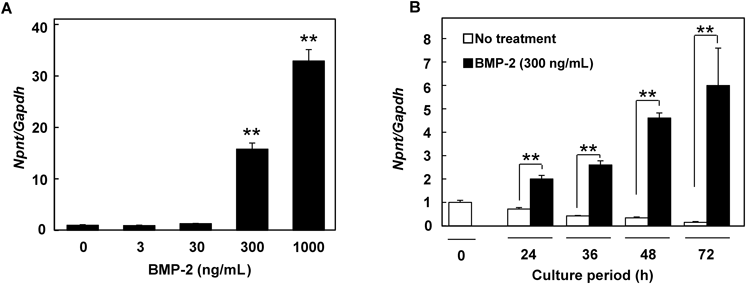
(A) Dose-dependent effects of BMP-2 on expression of Npnt mRNA in C2C12 cells after treatment with 0, 3, 30, 300, or 1000 ng/mL for 48 h. At higher concentrations, Npnt mRNA was remarkably up-regulated by BMP-2. Values are shown as the mean±S.D. of 3 samples folded over the group without treatment. ** p<0.01, relative to level in cells without treatment (ANOVA). (B) Time-dependent effects of BMP-2 on Npnt mRNA expression in C2C12 cells. After a 24-h pre-culture, cells were treated with 300 ng/mL of BMP-2, then harvested after 24, 36, 48, or 72 h. Real-time PCR analyses were performed using cDNA from each sample to examine the mRNA levels of Npnt and Gapdh. Values are shown as the mean±S.D. of 3 samples folded over the group taken at 0 h. ** p<0.01, relative to level in cells with no treatment at each time point (Student’s t-test).
C2C12 cells which are one of the mouse myoblast cell lines are used as a model for osteoblast differentiation induced by BMP-2. Analyses of ALP activity and MHC immunohistochemistry are revealed that C2C12 cells were differentiated into osteoblasts or myotubles using low FBS (0.5%) containing medium with or without BMP-2 (Fig. 1A). To examine their effect on Npnt mRNA expression, we exposed C2C12 cells to various BMPs for 48 h, then determined the level of Npnt mRNA using real-time PCR analysis (Fig. 1B). After separate treatments with BMP-2, -4, -6, and -7, each of which is known to have a strong osteogenic ability, the expression of Npnt mRNA was remarkably up-regulated. On the other hand, no increase was seen with BMP-3 treatment. It has been reported that, BMP-2, -4, -6, and -7 bind to the type I receptor ALK2/3 based on their affinity for BMP receptors,10) while BMP-3 is known as an agonist of activin receptor type 2b (Acvr2b).15) Moreover, BMP-3 has only a 50% identical amino acid sequence with other osteogenic BMPs16) and, in some cases, suppresses BMP functions.17) In consideration of these inherent differences, we believe that the present results are in agreement with previous reports. The expression level of Npnt protein was also increased in C2C12 cells after 48 h of incubation with BMP-2 (Fig. 1C).
In order to confirm that the observed up-regulation of Npnt mRNA expression was caused by the BMPs, we exposed C2C12 cells to various doses of BMP-2, the most widely used BMP to drive osteoblast differentiation, for different periods of time, and found that the expression was up-regulated in both dose- and time-dependent manners (Figs. 2A, B).
Based on those findings, we next utilized an ALK2/3 inhibitor (LDN193189) to determine the mechanism by which BMP-2 regulates the expression of Npnt mRNA in C2C12 cells (Fig. 3A). Since exposure to the ALK inhibitor canceled up-regulation by BMP-2, we speculated that the up-regulation of Npnt mRNA expression occurred via activation of the BMP receptor. To examine the effect of the ALK2/3 inhibitor, we performed Western blotting and detected phosphorylated Smad1 and Smad5, which were located in the downstream signaling pathway triggered by ALK2/3 receptors. As shown in Fig. 3B, the ALK2/3 inhibitor completely blocked phosphorylation of Smad1 and Smad5. Furthermore, we investigated whether the SMAD signaling pathway is activated on BMP-2 induced up-regulation of Npnt mRNA expression using siRNA of Smad4, a SMAD signaling factor that forms a complex with phosphorylated Smad1/5 and transmits BMP signals into the nucleus. Western blotting analysis showed that cells transfected with Smad4 siRNA had lower levels of Smad4 protein as compared to those transfected with control siRNA (Fig. 4A). Moreover, we transfected a construct containing a Smad4-responsive element (id1.0-luc vector) into Smad4 knockdown C2C12 cells14) and performed luciferase assay analysis (Fig. 4B). C2C12 cells transfected with the id1.0-luc vector demonstrated a high level of luciferase activity after BMP-2 treatment (control), while Smad4 knockdown cells showed only one-third of the activity of the control. Smad4 knockdown of C2C12 cells also resulted in a weaker up-regulation of Npnt mRNA expression by BMP-2 as compared to the control siRNA-transfected cells (Figs. 4C, D). Based on these results, we concluded that BMP-2 up-regulates Npnt mRNA expression in C2C12 cells via the BMP-SMAD signaling pathway.
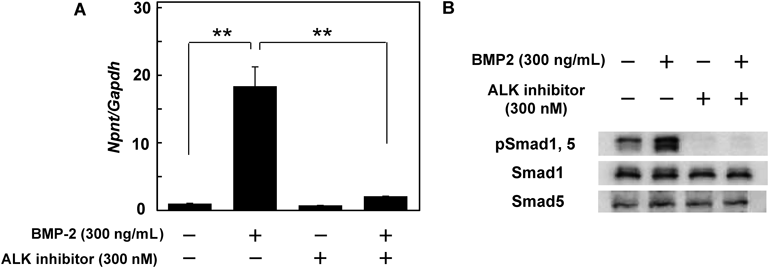
(A) The ALK2/3 inhibitor negated the up-regulation of Npnt mRNA expression by BMP-2. To block BMP-2 up-regulation, cells were treated with 300 ng/mL of BMP-2 and 300 nM of an ALK2/3 inhibitor for 48 h. Real-time PCR was performed using cDNA derived from total cellular RNA from each sample to determine the expression levels of Npnt and Gapdh mRNAs. Vales are shown as the mean±S.D. of 3 samples folded over the group without treatment. ** p<0.01, relative to level in cells without treatment (ANOVA). (B) ALK2/3 inhibitor blocked BMP-2-induced phosphorylation of Smad1 and Smad5 in C2C12 cells. After pre-treatment with 300 nM of the ALK2/3 inhibitor for 1 h, cells were exposed to medium containing 300 ng/mL of BMP-2 and 300 nM of the inhibitor, or to control medium without the inhibitor for 15 min. All proteins were extracted and used to determine the levels of phosphorylated and total Smad1 and Smad5.
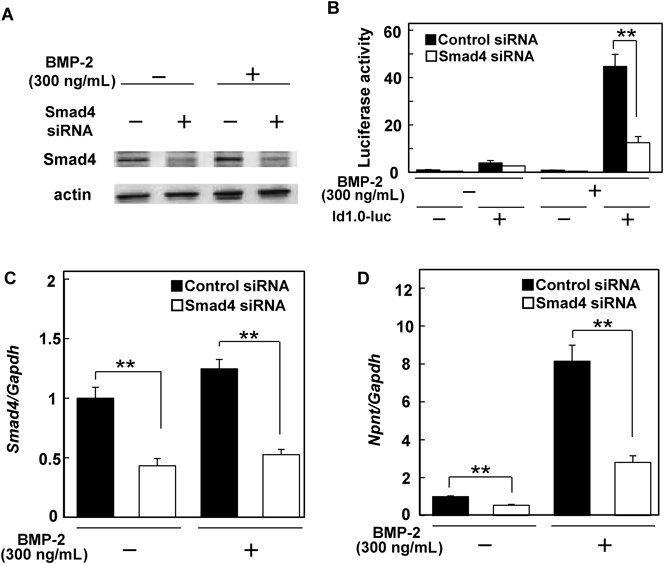
(A) Western blotting analysis of Smad4 in C2C12 cells transfected with or without Smad4 siRNA. After transfection of Smad4 siRNA for 48 h, cells were incubated with or without 300 ng/mL of BMP-2 for 48 h. Next, 5 µg of protein from each cell lysate was placed onto gels to detect Smad4 and actin. A Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific Inc., #23225) was used to measure (562 nm) the total protein concentration, which was compared to BSA protein used as the standard. (B) Luciferase assay analysis of Smad4 knockdown and id1.0-luc vector-transfected C2C12 cells. Values are shown as the mean±S.D. of 3 samples folded over the group transfected with the control siRNA and control vector. ** p<0.01, relative to level of cells transfected with control siRNA and control vector (ANOVA). (C) Smad4 was reduced in cells transfected with smad4 siRNA. Cells transfected with non-silencing siRNA were harvested as a control. Values are shown as the mean±S.D. of 6 samples folded over the group transfected with control siRNA without BMP-2 treatment. ** p<0.01, relative to level in cells transfected with control siRNA (Student’s t-test). (D) Both constitutive and up-regulated expressions of Npnt mRNA induced by BMP-2 were decreased in Smad4 siRNA-transfected cells. The expressions of Smad4, Npnt, and Gapdh were determined using real-time PCR analysis. Values are shown as the mean±S.D. of 6 samples folded over the group transfected with control siRNA without BMP-2 treatment. ** p<0.01, relative to level in cells transfected with control siRNA (Student’s t-test).
Our finding that up-regulation of Npnt by BMP-2 requires Smad4 implies the presence of a mechanism that regulates the expression of Npnt mRNA. Other recent studies have also indicated a competitive relationship between Smad2 and Smad3 or Smad4 involved in regulation of the expressions of some genes, such as hepatocyte growth factor (HGF).18–20) As for the expression of Npnt, Tsukasaki et al. reported its suppression by Smad2,21) while the present findings demonstrated its activation by Smad4. Together, these observations led us to speculate that the Npnt expression is also regulated by a transcription co-activator protein such as CBP/p300, and mediated by the competition between Smad2 and Smad4, though additional experiments are needed to more fully reveal the mechanism details.
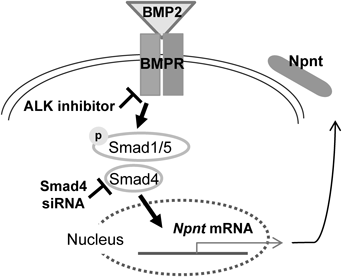
The receptor activated by BMP-2 phosphorylates Smad1/5, then phosphorylated Smad1/5 forms a complex with Smad4 to regulate the expressions of specific genes. The ALK2/3 inhibitor blocks phosphorylation of Smad1 and Smad5, while Smad4 siRNA reduces the up-regulation of Npnt mRNA expression induced by BMP-2.
The authors express their gratefulness to Dr. Kentaro Yoshimura for the valuable discussion. We also thank Dr. Satomi Nimura for guidance regarding the use of statistics software produced by SSRI for one-way ANOVA. This work was supported in part by the Project to Establish Strategic Research Center for Innovative Dentistry by the Ministry of Education, Culture, Sports, Science and Technology of Japan, and Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science.
The authors declare no conflict of interest.