2016 Volume 39 Issue 7 Pages 1144-1149
2016 Volume 39 Issue 7 Pages 1144-1149
In many parts of the world, the possession and cultivation of Cannabis sativa L. are restricted by law. As chemical or morphological analyses cannot identify the plant in some cases, a simple yet accurate DNA-based method for identifying C. sativa is desired. We have developed a loop-mediated isothermal amplification (LAMP) assay for the rapid identification of C. sativa. By optimizing the conditions for the LAMP reaction that targets a highly conserved region of tetrahydrocannabinolic acid (THCA) synthase gene, C. sativa was identified within 50 min at 60–66°C. The detection limit was the same as or higher than that of conventional PCR. The LAMP assay detected all 21 specimens of C. sativa, showing high specificity. Using a simple protocol, the identification of C. sativa could be accomplished within 90 min from sample treatment to detection without use of special equipment. A rapid, sensitive, highly specific, and convenient method for detecting and identifying C. sativa has been developed and is applicable to forensic investigations and industrial quality control.
Cannabis sativa L. (Family Cannabaceae) has been cultivated for thousands of years for use as food, medicine, and fiber.1) C. sativa seeds are a good source of nutrition because of their high quality protein and lipid.2) Also called mashinin, the seeds are used as a laxative in Kanpo medicine. Its fiber is utilized in the manufacture of rope, paper, and fabric. However, despite these notable characteristics, the cultivation and possession of C. sativa are restricted by law in many parts of the world because C. sativa possesses strong psychoactive effects.3) C. sativa tops and leaves contain cannabinoids, which are the characteristic chemical compounds in C. sativa. More than 70 cannabinoids, including tetrahydrocannabinolic acid (THCA), have been isolated from C. sativa.4–6) Tetrahydrocannabinol (THC), the decarboxylated form of THCA, is the primary cannabinoid responsible for the strong psychoactivite effects.1,7)
The identification of C. sativa species is crucial for forensic investigation and industrial quality control. C. sativa can be discriminated from other species by morphological analysis using cystolithic hairs on the leaves or chemical analyses using HPLC, liquid chromatography-mass spectrometry (LC-MS), or GC-MS.8–13) However, it is difficult to distinguish C. sativa samples as they lack distinct morphological or chemical characteristics. In those cases, DNA analyses are used for species identification. It has been reported that DNA-based assays using PCR or real-time quantitative PCR are able to detect C. sativa, but those assays require long hours for DNA extraction and special equipment, such as a thermal cycler.14–17)
Loop-mediated isothermal amplification (LAMP) is a nucleotide acid amplification method that features high sensitivity, specificity, and rapidity under isothermal conditions.18) LAMP requires a set of four primers to react with six distinct regions in the target. The reaction is accelerated by adding two loop primers and can be visually detected by the white turbidity that results from magnesium pyrophosphate accumulation during amplification.19,20) It does not need any special devices or experienced technicians; thus, it is widely used for various purposes, such as detecting viruses and medicinal herbs.21–24)
One of the most species-specific genes in C. sativa is the THCA synthase gene that consists of a 1632 bp open reading frame. THCA synthase gene was cloned and polymorphism analysis revealed that 63 nucleotide substitutions divided the species into two chemotypes: the THCA-rich type (drug type), such as marijuana, and the THCA-poor type (fiber type), such as hemp.25) It has been suggested that 37 amino acid substitutions corresponding to the polymorphism determine the ability of THCA synthesis.25–27) THCA synthase gene does not have high homology with any genes of other species. In this study, we have developed a novel LAMP assay for detecting C. sativa by targeting the conserved region of THCA synthase gene. This method is rapid, sensitive, highly specific, and convenient.
We used 20 specimens of C. sativa from the Medical Plant Garden, Tokyo Metropolitan Institute of Public Health, one specimen of C. sativa from Hokuriku University, which has been granted permission to study C. sativa by Ishikawa Prefecture, and seven species, namely, Humulus lupulus, Humulus japonicus, Cudrania tricuspidata, Paeonia lactiflora, Angelica acutiloba, Matricaria recutita, and Glycyrrhiza glabra, from the Medicinal Plant Garden, Kanazawa University. The specimens of C. sativa had been cultivated in each institute, but the origin of genetic background is unknown. All specimens were deposited in the herbarium of Kanazawa University and the Forensic Science Laboratory in Ishikawa Prefecture. Total genomic DNA was extracted from each dried leaf using a DNeasy Plant Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. Each DNA concentration was measured at 260 nm and adjusted to the final concentration of 5 ng/µL with sterile water for use.
Primer DesignLAMP primers were designed on the basis of fiber-type THCA synthase gene (DDBJ/EMBL/GenBank database accession No. AB212830). Regarding polymorphism, THCA synthase gene is one of the most analyzed genes in C. sativa. Drug-type (accession No. AB212834) and fiber-type (accession No. AB212830) THCA synthase genes were aligned and the conserved regions were selected to design LAMP primers using PrimerExplorer V3 and V4 (https://primerexplorer.jp). Two outer primers, one forward primer (FIP), and one backward inner primer (BIP) were designed for the LAMP reaction. A loop primer (LF) was designed to accelerate the reaction. The primer sequences are indicated in Fig. 1.
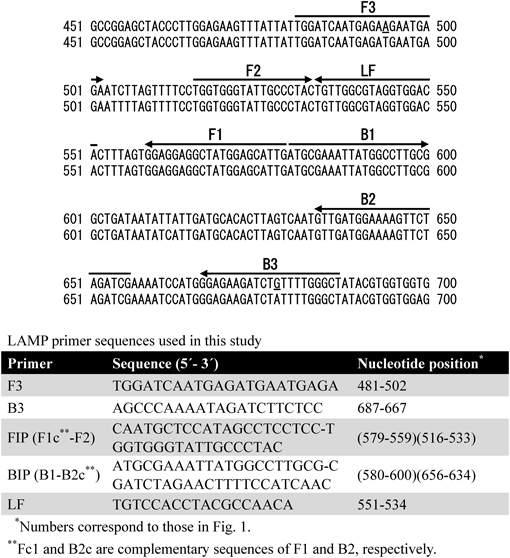
Nucleotide sequence of THCA synthase gene. The upper array is the drug-type sequence (accession No. AB212834) and the lower array is the fiber-type sequence (accession No. AB212830). The sequences of the primer binding sites are indicated by arrows. Nucleotide differences in the used primer sequences are underlined.
The LAMP reaction was performed with a Loopamp DNA Amplification Kit (Eiken Chemical Co., Ltd., Tokyo, Japan) according to the manufacturer’s protocol with minor modification. Briefly, a 25 µL mixture containing 12.5 µL of 2×Reaction mix, 8 units of Bst DNA polymerase, 0.5 µL of Loopamp Fluorescent Detection Reagent (Eiken Chemical Co., Ltd.), 0.2 µM each of F3 and B3 primers, 1.6 µM each of FIP and BIP primers, 0.8 µM of LF primer, and 1 µL of each genomic DNA as template was incubated at 60, 63, 66, and 69°C for 90 or 150 min and then heated at 80°C for 5 min by a Smart Cycler (Cepheid). The fluorescence produced when calcein formed a chelate with magnesium ion was observed at wavelengths between 565 and 590 nm. The baseline and the threshold cycle (Ct) were automatically determined by Smart Cycler. Ct was converted into the initial rise time.
Sensitivity of LAMP AssayLAMP assay and conventional PCR were performed with serial 10-fold dilutions of genomic DNA of Afghani special from 10 to 0.001 ng as template. Conventional PCR was conducted in a final volume of 25 µL containing 1×PCR buffer (TaKaRa, Japan), 0.2 mM of each deoxyribonucleotide triphosphate (dNTP) (TaKaRa), 0.5 µM each of F3 and B3 primers, 0.625 unit of Ex Taq DNA polymerase (TaKaRa), and 1 µL of each genomic DNA. PCR conditions were as follows: initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s, with a final extension at 72°C for 5 min, followed by a 4°C hold. The PCR products were electrophoresed on 2% agarose gels with ethidium bromide staining.
Specificity of LAMP AssayLAMP specificity was tested with the DNA samples extracted from the 21 specimens of C. sativa as positive control and from seven species, including H. lupulus and H. japonicus, which are closely related to C. sativa, as negative control.
Sequencing Analysis and CloningTHCA synthase gene was amplified by PCR using forward primer (5′→3′; GCG GAT CCA TGA ATT GCT CAG CAT TTT CCT TTT) and reverse primer (5′→3′; GCC TGC AGT CTA TTT AAA GAT AAT TAA TGA TGA TGC GGT GG) of reported oligonucleotide primers with minor modification.25) The PCR products were treated with ExoSAP-IT (Affymetrix) to remove unconsumed dNTPs and excess primers. The purified PCR products were sequenced with a BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems) and a 3130xl Genetic Analyzer (Applied Biosystems) according to the manufacturer’s instructions.
Two types of THCA synthase genes were cloned as follows. PCR products of THCA synthase gene obtained from C. sativa (Big Bud) were digested with BamHI and PstI, ligated into pBluescript vector, and transformed in E. coli. Sequence analysis showed that each clone of THCA synthase gene had the sequence of drug type (accession No. LC120319) or fiber type (accession No. LC120320) in the LAMP targeting region and these sequences were registered by authors. Constructed plasmids were diluted to 0.01 fmol/µL with sterile water for use.
Simple Sample Preparation for LAMP AssayA piece of dried leaf (approximately 3 mg) was added to 100 µL of lysis buffer consisting of phosphate buffered saline (PBS) with a final concentration of 5% Tween 20 and 1 µg/µL proteinase K (Qiagen). The lysis sample was heated at 56°C for 5 min and then at 95°C for 5 min, and centrifuged. As DNA template, 1 µL of the supernatant was used for the LAMP assay.
We aligned THCA synthase gene and selected the conserved regions in the THCA synthase gene for designing LAMP primers (Fig. 1). LAMP primers consist of two outer primers (F3, B3), two inner primers (FIP, BIP), and a loop primer (LF). The forward inner primer (FIP) consists of F1c and F2, and the backward inner primer (BIP) consists of B1 and B2c. F1c and B2c are complementary sequences of F1 and B1, respectively. Each binding site of F3 and B3 contains a single nucleotide polymorphism between the drug-type and fiber-type THCA synthase genes. As each single nucleotide polymorphism is located in the middle of the primer sequence, it was probable that the mutation did not influence the LAMP reaction. In addition, to reduce the reaction time, an LF primer was designed.
Optimization of LAMP Reaction ConditionsThe LAMP assay using a set of primers for detecting C. sativa was performed. The fluorescence derived from the calcein-magnesium chelate was measured at wavelengths between 565 and 590 nm using Smart Cycler28) (Fig. 2A). With C. sativa DNA as template, the initial rise time was 92.9±2.6 min without LF. When LF was added, the initial rise time was 39.3±1.9 min, more than 50 min shorter than the time without LF. Moreover, agarose gel electrophoresis revealed a typical ladder-like pattern in a positive sample (Fig. 2B). As LAMP assays have 100 to 1000-fold amplification efficiency compared with PCR in general, cross-contamination caused by opening tubes after LAMP reaction poses a serious problem. To avoid cross-contamination, the electrophoresis of LAMP products was performed in a physically separated room. The amplification product was easily observed as green fluorescence under natural light (Fig. 2C). Under ultraviolet irradiation, it was also detected as the fluorescence of the calcein-magnesium chelate (Fig. 2D).

LAMP reactions were monitored by Smart Cycler (A). Fluorescence signal (y-axis) was observed on a channel that detects wavelengths between 565 and 590 nm in real time. LAMP reactions with a loop primer (LF (+)) or without a loop primer (LF (−)) were performed. Positive reactions were visualized by agarose gel electrophoresis (B), under natural light (C), and by UV irradiation (D).
To determine the optimum reaction temperature, the LAMP assay was conducted at 60, 63, 66, and 69°C (Fig. 3). The amplifications occurred at 44.6±2.9, 39.3±1.9, and 44.3±4.3 min at 60, 63, and 66°C, respectively, but not at 69°C. There was no non-specific amplification occurring at 90 min at any temperature. Because the initial rise time at 63°C was shorter than those at 60 and 66°C, we determined that the optimum temperature for the assay was 63°C whereas the amplification could be detected at a broad range of temperatures between 60 and 66°C within 50 min. These results indicated that the LAMP assay does not require a precise temperature control system, such as a thermal cycler, and a simple device, such as a water bath or a heating block, is sufficient for the assay.

The LAMP assay was conducted at 60, 63, 66, and 69°C. Amplification was detected at 60, 63, and 66°C, but not at 69°C. Non-specific amplification did not occur at any temperature.
To evaluate the sensitivity, serial 10-fold dilutions of genomic DNA (10 to 0.001 ng) were used as DNA template for the LAMP assay and PCR. The fluorescence intensity was enhanced with the DNA template of 10, 1, and 0.1 ng, and the initial rise time was 37.8±2.0, 41.8±1.4, and 52.2±7.1 min, respectively. No amplification was observed with the 0.01 ng DNA template (Fig. 4A). For comparison with conventional PCR, PCR using F3 and B3 was carried out. By agarose gel electrophoresis, amplification of THCA synthase gene was detected from 0.1 to 10 ng, but the PCR product of the 0.1 ng DNA template was not clearly observed (Fig. 4B). These results demonstrated that the sensitivity of the LAMP assay was the same as or higher than that of PCR and the minimum amount of DNA template necessary for the LAMP assay was 0.1 ng. Furthermore, our LAMP assay showed higher sensitivity than the previously reported assay that required 5 ng of DNA template to discriminate species for various purposes.15)

Serial 10-fold dilutions of genomic DNA (10 to 0.001 ng) were used as DNA template for the LAMP assay (A) and PCR (B). Sensitivity of the LAMP assay was the same as or higher than that of conventional PCR.
The specificity of the LAMP assay for 21 specimens of C. sativa and seven other species, including H. lupulus and H. japonicus, was tested. All specimens of Cannabis spp. were detected in the LAMP reaction within 45 min (Table 1). No other species, including closely related species, such as H. lupulus and H. japonicus, were amplified within 90 min. Regarding the reaction time, the initial rise times of the 21 specimens varied from 31.5±0.7 min (No. 14; PURPLE) to 39.5±0.4 min (No. 10; Leda Uno). To investigate reaction time differences among the specimens, we performed sequence analyses of the LAMP targeting region in THCA synthase gene. The sequence analyses revealed that 13 specimens had only the drug-type sequence and 8 specimens had both drug-type and fiber-type sequences. The average initial rise time of the group having both drug-type and fiber-type THCA synthase genes was 33.2±1.1 min, shorter than that of the group having only drug-type THCA synthase gene (37.8±1.8 min). Because the LAMP primers were designed on the basis of the fiber-type sequence, it is probable that misannealing of the drug-type THCA synthase gene increased the reaction time. However, these slight differences in the reaction time did not pose any problems in C. sativa detection because several factors, such as DNA concentration, DNA quality, reagent lot, and devices, could influence the reaction time. In this study, there were no specimens having only fiber-type THCA synthase gene. To test the LAMP reactivity for the fiber-type THCA synthase gene, we conducted the LAMP assay using the cloned THCA synthase gene as template. As shown in Fig. 5, the LAMP assay was able to detect each type of THCA synthase gene. The initial rise times of drug- and fiber-type THCA synthase genes when 0.01 fmol plasmid DNA was used were 33.8±1.3 and 31.0±1.4 min, respectively. These results indicated that the LAMP primer set is highly specific to both gene types. Thus, C. sativa having only fiber-type THCA synthase gene can be detected with this method. Accordingly, the results showed that this method has high species specificity for C. sativa.
| No. | Product name (Species) | THCA synthase gene type | Initial rise time |
|---|---|---|---|
| 1 | Afghani special | Drug | 39.3±1.9 |
| 2 | Bahia Black Head | Drug | 37.2±0.6 |
| 3 | Big Bud | Drug/Fiber | 32.5±0.4 |
| 4 | CAL. ORANGE | Drug/Fiber | 31.8±0.4 |
| 5 | Crystal Paradise | Drug | 38.6±0.7 |
| 6 | Durban Poison | Drug/Fiber | 33.6±0.4 |
| 7 | Early Bud | Drug | 37.4±0.5 |
| 8 | Haze Special | Drug | 38.8±1.2 |
| 9 | ISIS | Drug | 37.6±1.1 |
| 10 | Leda Uno | Drug | 39.5±0.4 |
| 11 | Mango | Drug | 35.4±0.2 |
| 12 | Northern Light | Drug/Fiber | 34.2±0.7 |
| 13 | Northern Light Special | Drug | 37.4±0.7 |
| 14 | PURPLE | Drug/Fiber | 31.5±0.7 |
| 15 | PURPLE STAR | Drug/Fiber | 34.0±0.2 |
| 16 | SACRA FRASCA | Drug | 38.3±0.5 |
| 17 | SHAMAN | Drug/Fiber | 33.2±0.6 |
| 18 | SKUNK PASSION | Drug | 37.4±0.7 |
| 19 | TWILIGHT | Drug | 37.7±0.9 |
| 20 | VOODOO | Drug | 34.2±0.4 |
| 21 | Unknown | Drug/Fiber | 34.4±0.5 |
| N1 | (Humulus lupulus) | — | Not detected |
| N2 | (Humulus japonicus) | — | Not detected |
| N3 | (Cudrania tricuspidata) | — | Not detected |
| N4 | (Paeonia lactiflora) | — | Not detected |
| N5 | (Angelica acutiloba) | — | Not detected |
| N6 | (Matricaria recutita) | — | Not detected |
| N7 | (Glycyrrhiza glabra L.) | — | Not detected |

The reactivity of constructed plasmid containing drug-type THCA synthase gene (accession No. LC120319) or fiber-type THCA synthase genes (accession No. LC120320) was tested by LAMP assay. Amplifications were noted with both types of DNA templates, whereas no amplification was noted with empty vector (pBluescript vector).
Because some inhibitory compounds exist in plant tissues, such as polysaccharides or secondary metabolites, direct amplification of unpurified DNA seems to be difficult for enzymatic reactions in general. However, as Bst polymerase, which is used in the LAMP assay, is robust to the inhibitors, some protocols that require no DNA extraction have been reported.29) To simplify the DNA extraction procedure, a simple method for sample preparation was evaluated for the LAMP assay.30,31) Supernatants of heat-treated samples following proteinase treatment were used as DNA template. As indicated in Fig. 6, the purified DNA template and the supernatant of C. sativa leaf (No. 18; SKUNK PASSION) showed fluorescence amplification at 37.4±0.7 and 45.8±2.2 min, respectively, whereas the supernatants of leaves of the other species showed no amplification. The results indicate that identifying C. sativa leaf can be completed within 90 min from sample preparation to detection.

Leaf samples were treated with proteinase, heated, and centrifuged. The supernatant of C. sativa leaf showed fluorescence amplification whereas the supernatants of leaves of other species showed no amplification (A). A typical LAMP product (ladder-like pattern) was detected from C. sativa leaf by agarose gel electrophoresis (B).
We have developed a LAMP assay to identify C. sativa using LAMP primers with conserved THCA synthase gene. The LAMP assay is convenient and its sensitivity is the same as or higher than those of conventional DNA-based methods.The LAMP reaction can be performed under isothermal conditions at 60–66°C without the need for special equipment. Moreover, the simple procedure using unpurified DNA template can be accomplished within 90 min from pretreatment to detection. These characteristics indicate that the LAMP assay is suitable for on-site detection or less well-equipped laboratories. This rapid, sensitive, highly specific, and convenient method for detecting and identifying C. sativa may be used as a reliable tool for forensic investigation and industrial quality control.
This research was partly supported by a Grant from the Research Foundation for Safe Society. We thank our colleagues for the support extended in the course of this study.
The authors declare no conflict of interest.