2016 Volume 39 Issue 9 Pages 1432-1436
2016 Volume 39 Issue 9 Pages 1432-1436
Dextrorphan, an active metabolite of the antitussive dextromethorphan, has been shown to be subjected to sulfation by several zebrafish cytosolic sulfotransferases (SULTs). We were interested in finding out which of the human SULT(s) is(are) capable of catalyzing the sulfation of dextrorphan, and to verify whether sulfation of dextrorphan may occur in cultured human cells and human organ cytosols. Data from the enzymatic assays showed that, of all thirteen known human SULTs, SULT1A3 displayed the strongest dextrorphan-sulfating activity. Cell culture experiments using HepG2 human hepatoma cells and Caco-2 human colon carcinoma cells incubated with [35S]sulfate together with varying concentrations of dextrorphan revealed indeed the production and release of [35S]sulfated dextrorphan in a concentration-dependent manner. Additionally, significant dextrorphan-sulfating activity was detected in human liver, small intestine and lung cytosols. Taken together, these results provided a biochemical basis for the sulfation of dextrorphan in humans.
Dextromethorphan is an antitussive commonly used for treating cough associated with upper respiratory tract infections such as common cold and influenza.1) Upon oral administration, dextromethorphan is absorbed from the gastrointestinal tract, reaching peak concentrations in circulation in approximately 2.5 h.2) In the liver, dextromethorphan is subjected to O-demethylation under the action of cytochrome P450 2D6 (CYP2D6), generating dextrorphan.3,4) Dextrorphan has been shown to act as an antagonist of the excitatory neurotransmitter N-methyl-D-aspartate (NMDA),5) being more potent than dextromethorphan.6,7) It has thus been suggested that the effect of dextromethorphan may be due to conversion to dextrorphan.8) An interesting issue is whether dextrorphan is subjected to further metabolism in the body.
Sulfate conjugation plays a critical role in the biotransformation of many xenobiotics, including drugs, in vertebrates.9–11) In sulfate conjugation reactions, the cytosolic sulfotransferase (SULT) enzymes mediate the transfer of a sulfonate group from the donor compound, 3′-phosphoadenosine 5′-phosphosulfate (PAPS), to amino or hydroxyl group of the substrate compounds.12) Sulfate conjugation generally leads to the inactivation of the substrate compounds and concomitantly increase their water-solubility, thereby facilitating their excretion from the body.13) In humans, thirteen distinct SULTs have been identified and classified into four SULT families, SULT1, SULT2, SULT4, and SULT6.14,15) In an earlier study, several zebrafish SULTs were found to be capable of sulfating dextrorphan.16) Whether dextrorphan can be sulfated by any of the human SULTs, however, remains unknown.
In this study, we examined all known human SULTs for potential sulfating activity towards dextrorphan. Furthermore, possible sulfation of dextrorphan in cultured human cells and by human organ specimens was investigated.
Dextrorphan tartrate and levorphanol tartrate were obtained from Cerilliant Corporation (Round Rock, TX, U.S.A.). Dopamine, p-nitrophenol, adenosine 5′-triphosphate, dithiothreitol, N-2-hydroxylpiperazine-N′-2-ethanesulfonic acid (HEPES), minimum essential medium (MEM), and fetal bovine serum (FBS) were products of Sigma Chemical Company (St. Louis, MO, U.S.A.). Sep-Pak C18 cartridges were products of Waters Corporation (U.S.A.). Sodium [35S]sulfate was purchased from American Radiolabeled Chemicals (St. Louis, MO, U.S.A.). Recombinant human SULTs were expressed using Escherichia coli BL21 host cells and isolated as previously described.17–21) PAP[35S] was enzymatically synthesized from adenosine 5′-triphosphate and carrier-free [35S]sulfate using human adenosine 5′-triphosphate sulfurylase/adenosine 5′-phosphosulfate kinase.22) Caco-2 human intestinal epithelial cell line (ATC C HTB-37) and HepG2 human hepatoma cell line (ATC C HB-8065) were obtained from American Type Culture Collection (Manassas, VA, U.S.A.). Human intestine, kidney, liver, and lung cytosols were from XenoTech, LLC (Lenexa, KS, U.S.A.). Cellulose TLC plates were products of EMD Millipore Corporation (Billerica, MA, U.S.A.). Ecolume scintillation cocktail was from MP Biomedicals, LLC (Irvine, CA, U.S.A.).
Enzyme AssayThe dextrorphan-sulfating activity of the recombinant human SULTs was determined by enzymatic assay using radioactive PAP[35S] as the sulfate donor. A typical assay mixture, with a final volume of 20 µL, contained 14 µM PAP[35S], 1 mM dithiothreitol, 50 mM of HEPES buffer (pH 7.0), and 50 µM of dextrorphan. The assay was initiated by adding the tested SULT enzyme, allowed to continue for 5 min at 37°C and stopped by heating the assay mixture-containing tube on a 100°C dry block heater for 3 min. Precipitates formed during the heat inactivation were removed by centrifugation for 3 min at 13000×g, and the cleared supernatant was used for the TLC analysis for [35S]sulfated dextrorphan. The solvent system used was n-butanol–isopropanol–88% formic acid–water (2 : 1 : 1 : 2, v/v). At the end of TLC, the plate was dried in a hood, followed by autoradiography using an X-ray film. The radioactive spot corresponding to [35S]sulfated dextrorphan was located on the TLC plate, and cut out and eluted in water (0.5 mL) in a 5 mL glass vial. Subsequently, 4.5 mL of Ecolume scintillation cocktail was added, mixed thoroughly, and the [35S]radioactivity therein was quantified by liquid scintillation. In a concentration-dependent experiment, enzymatic assays were carried out using varying concentrations of dextrorphan as substrates. The reaction conditions and the procedure for the analysis of [35S]sulfated dextrorphan were the same as those described above. To analyze the dextrorphan-sulfating activity of human organ cytosols, the assay mixture was fortified with 50 mM NaF (a phosphatase inhibitor). The reaction was initiated by adding the tested human cytosol and allowed to continue for 30 min, followed by the TLC analysis for [35S]sulfated dextrorphan as described above.
Mass Spectrometry AnalysisTo prepare sulfated dextrorphan for structural analysis using mass spectrometry, enzymatic sulfation of dextrorphan by SULT1A3 was performed using non-radioactive PAPS as the sulfate donor based on the above-mentioned procedure. The final reaction mixture was applied to a Waters Sep-Pak C18 cartridge. The bound dextrorphan sulfate was eluted using water–methanol (2 : 3, v/v), and the eluate was dried using a SpeedVac concentrator. The dry residue was reconstituted using methanol and analyzed using a Shimadzu Nexera XR HPLC system and LCMS-8050 triple quadrupole mass spectrometer with negative ion mode. Ten microliters of sample was injected with the mobile phase consisting of water–methanol (1 : 1, v/v) plus 0.1% formic acid at a flow rate of 0.4 mL/min into a capillary without separation column. Mass spectrometry conditions employed were: Nebulizing Gas Flow, 2.0 L/min; Drying Gas Flow, 10.0 L/min; Heating Gas Flow, 10.0 L/min; ESI Interface Voltage, 3.0 kV; Interface Temperature, 300°C; DL Temperature, 250°C; IG Vacuum, 0.02 Pa; PG Vacuum, 92 Pa; and CID Gas, 270 kPa.
Metabolic Labeling Study Using Cultured Human CellsCaco-2 cells and HepG2 cells were routinely cultured in MEM plus 10% FBS, penicillin G (30 µg/mL), and streptomycin sulfate (50 µg/mL). Confluent cells grown in separate wells of a 24-well culture plate were first pre-incubated in sulfate-free MEM plus 10% dialyzed FBS for 4 h and then incubated in 0.25 mL aliquots of the same medium containing 0.3 mCi/mL [35S]sulfate plus varying concentrations (0, 5, 10, 25, 50, 100 µM) of dextrorphan. Following an 18-h labeling, the media were collected, centrifuged, and analyzed for the presence of [35S]sulfated dextrorphan based on aforementioned TLC procedure.
Previous studies using the zebrafish SULTs revealed that dextrorphan may be metabolized through sulfation.16) This study aimed to investigate whether and which of the human SULT(s) is(are) capable of catalyzing the sulfation of dextrorphan. Moreover, possible sulfation of dextrorphan in cultured human cells and by human organ cytosols was evaluated.
Identification of Human SULT(s) Capable of Sulfating DextrorphanA panel of thirteen human SULTs was analyzed for sulfating activity with dextrorphan as a substrate. Results obtained showed that nine (SULT1A2, SULT1B1, SULT1C2, SULT1C3, SULT1E1, SULT2B1a, SULT2B1b, SULT4A1, SULT6B1) of the thirteen human SULTs displayed no detectable activities. Of the other four SULTs (SULT1A1, SULT1A3, SULT1C4, SULT2A1), SULT1A3 exhibited much stronger activities than the other three towards dextrorphan (Table 1). Based on these results, SULT1A3 is likely the major enzyme responsible for the sulfation of dextrorphan in human body. Since substrate and/or product inhibition may occur for SULTs,9,23) a concentration-dependent experiment was performed using different concentrations of dextrorphan as substrates for all four dextrorphan-sulfating SULTs. As shown in Fig. 1, no significant substrate or product inhibition was observed over the tested dextrorphan concentration ranges (up to 1 mM for SULT1A3 and SULT2A1, and up to 2 mM for SULT1A1 and SULT1C4). As shown in Table 1, in addition to dextrorphan, SULT1A3 was also capable of mediating the sulfation of dopamine (a physiological substrate), p-nitrophenol (a prototype substrate for phenol sulfotransferases), and levorphanol (a levorotatory enantiomer of dextrorphan). It is interesting to note that the sulfating activity of SULT1A3 with dextrorphan was more than 38 times higher than that with levorphanol, whereas the other three SULTs did not show similar stereoselectivity for dextrorphan and levorphanol. The sulfated product of dextrorphan generated by SULT1A3 was purified by solid-phase extraction using a Sep-Pak C18 reverse-phase cartridge and further analyzed using mass spectrometry. As shown in Fig. 2A, a prominent ion with m/z of 336, equivalent to a deprotonated mono-sulfated dextrorphan ion, was detected. This prominent ion produced a sulfonate ion with m/z of 80 and a de-sulfonated ion with m/z of 256, which is the dextrorphan ion (Fig. 2B). These results clearly indicated that the sulfation as mediated by SULT1A3 took place at 3-hydroxyl group of the aromatic ring structure of dextrorphan. Previous studies have demonstrated the expression of SULT1A3 in certain human organs including the brain, gastrointestinal tract, kidney, liver, and lung.24,25) It is possible that in these organs, SULT1A3 may serve to sulfate dextrorphan, a product generated following the O-demethylation of dextromethorphan by CYP2D6.3,4)
| Specific activity (nmol/min/mg) | ||||
|---|---|---|---|---|
| SULT1A1 | SULT1A3 | SULT1C4 | SULT2A1 | |
| Dextrorphan | 0.01±0.01 | 1.94±0.08 | 0.02±0.01 | 0.08±0.01 |
| Levorphanol | N.D. | 0.05±0.01 | 0.01±0.01 | 0.03±0.01 |
| Dopamine | 3.24±0.15 | 78.11±2.22 | 0.43±0.02 | N.D. |
| p-Nitrophenol | 34.52±1.27 | 2.93±0.16 | 81.20±1.67 | 0.07±0.01 |
a) Specific activity corresponds to nmol substrate sulfated/min/mg purified enzyme. Results shown represent the mean±standard deviation derived from three separate assays.

The concentrations of dextrorphan tested were 25, 50, 100, 200, 300, 400, 500, 600, 700, 800, 900, and 1000 µM for SULT1A3 and SULT2A1, and 100, 250, 500, 750, 1000, 1250, 1500, 1750, and 2000 µM for SULT1A1 and SULT1C4.
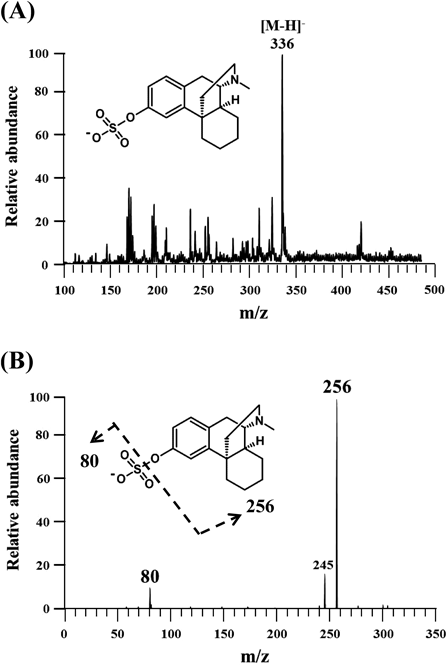
(A) Full mass spectrum of sulfated dextrorphan isolated using a Sep-Pak C18 cartridge. (B) MS2 spectrum of 336 m/z.
It is interesting to note that while all four human dextrorphan-sulfating SULTs belong to the SULT1 family, the two zebrafish SULTs previously shown to be capable of sulfating dextrorphan are members of the SULT3 family.16) Of the two zebrafish dextrorphan-sulfating SULTs, SULT3 ST1 displayed a specific activity of 0.89 nmol/min/mg, whereas SULT3 ST3 showed a specific activity of 0.10 nmol/min/mg. In terms of the dextrorphan-sulfating activity, zebrafish SULT3 ST1 appeared to resemble more closely the major human dextrorphan-sulfating SULT, SULT1A3 (cf. Table 1). Amino acid sequence analysis, however, failed to reveal a particularly high sequence homology between human SULT1A3 and zebrafish SULT3 ST1, as compared with other zebrafish SULTs (data not shown). It thus seems that, in different species, the sulfation of dextrorphan may be mediated by SULT members that bear no direct orthologous phylogenetic relationships.
Production and Release of [35S]Sulfated Dextrorphan by Cultured Caco-2 Cells and HepG2 Cells Incubated in Media Containing [35S]Sulfate and Varying Concentrations of DextrorphanFollowing the identification of human SULTs capable of sulfating dextrorphan, we were interested in finding out whether sulfation of dextrorphan may indeed occur in human cells. Cultured Caco-2 cells and HepG2 cells were used in this study. Previous studies have revealed the expression of several SULT enzymes, including SULT1A1, SULT1A2, SULT1A3, SULT1B1, SULT1C2, SULT1C4, and SULT2A1, in Caco-2 human intestinal epithelial cells.26) HepG2 human hepatoma cells, on the other hand, have been shown to express SULT1A1, SULT1A2, SULT1A3, SULT1E1, and SULT2A1.27,28) Confluent Caco-2 cells and HepG2 cells grown to confluence in different wells of a 24-well plate were incubated in sulfate-free medium containing [35S]sulfate and varying concentrations of dextrorphan. Following an 18-h incubation, the spent media collected were analyzed by the TLC procedure. As shown in Fig. 3, a significant amount of [35S]sulfated dextrorphan was detected in spent labeling medium containing as low as 5 µM of dextrorphan, which increased proportionately with increasing concentrations of dextrorphan added to the labeling media. These results showed clearly that both Caco-2 cells and HepG2 cells were capable of metabolizing dextrorphan by sulfation. As mentioned above, both HepG2 cells and Caco-2 cells have been reported to express SULT1A3, as well as SULT1A1 and SULT2A1,26–28) that are capable of sulfating dextrorphan (cf. Table 1).

Confluent Caco-2 cells and HepG2 cells were labeled with [35S]sulfate in the presence of varying concentrations (0, 5, 10, 25, 50, 100 µM) of dextrorphan. Following an 18-h incubation, spent media were collected and analyzed by TLC. The figure shows the autoradiograph taken from the TLC plate following the analysis. Lane E corresponds to [35S]sulfated dextrorphan produced enzymatically under the action of SULT1A3.
To investigate further whether sulfation of dextrorphan may occur in human organs, enzymatic assays were performed using cytosol fractions prepared from human lung, liver, kidney or intestine. Table 2 shows the activity data obtained from these assays. Of the four human organ cytosols tested, the cytosol prepared from human intestine exhibited the highest dextrorphan-sulfating activity, followed by the cytosols prepared from human liver and lung. Whereas the cytosol prepared from kidney showed no detectable dextrorphan-sulfating activity. These results implied that intestine and liver are likely major human organs involved in the metabolism of dextrorphan through sulfation. It should be pointed out that although SULT1A3, the major dextrorphan-sulfating SULT, is known to be present at much higher level in the intestine than in the liver, the level of dextrorphan-sulfating activity detected for the liver sample amounted to more than 50% of that detected for the small intestine sample (Table 2). It is possible that the still considerable dextrorphan-sulfating activity detected for the liver sample might have been, in part, due to the presence of other dextrorphan-sulfating SULTs (cf. Table 1), particularly SULT1A1 and SULT2A1, that are known to be expressed in the liver.29,30)
| Specific activity (pmol/min/mg protein) | |||
|---|---|---|---|
| Lung | Liver | Kidney | Small intestine |
| 0.15±0.05 | 2.35±0.12 | Not detected | 4.49±0.19 |
a) Specific activity corresponds to pmol substrate sulfated/min/mg protein. Results shown represent the mean±standard deviation derived from three separate assays. The concentration of the substrate used in the assay mixture was 50 µM.
To sum up, the present study revealed SULT1A3 as the major human SULT enzyme capable of sulfating dextrorphan. Cell culture experiments revealed clearly the occurrence of the sulfation of dextrorphan in human cells. Moreover, human intestine, liver, and lung cytosols were shown to be capable of mediating the sulfation of dextrorphan. Collectively, the results derived from the present study provided useful information relevant to the biochemical basis for the metabolism of dextrorphan through sulfation in vitro and in vivo.
This work was supported in part by a Grant from National Institutes of Health (Grant No. R03HD071146). Mass spectrometry was performed using instruments located in the Shimadzu Laboratory for Pharmaceutical Research Excellence in the College of Pharmacy and Pharmaceutical Sciences at the University of Toledo Health Science Campus.
The authors declare no conflict of interest.