2018 Volume 41 Issue 10 Pages 1561-1566
2018 Volume 41 Issue 10 Pages 1561-1566
Dietary taurine deficiency results in dilated cardiomyopathy in cats while in mice taurine deficiency produced by knocking out the taurine transporter (TauT) gene leads to a reduction in cardiac function with advancing age. The present study elucidated the involvement of cardiac fibrosis in the aging-dependent cardiac disorder of the TauT-knockout (TauTKO) mouse. Old (18–24-month-old) TauTKO mice, but not young (3–5-month-old) mice, exhibit cardiac fibrosis. Transcriptome microarray analysis revealed an increase in pro-fibrotic genes, such as S100A4, ACTA2 and CTGF, in both young and old TauTKO hearts. Based on transcriptome-pathway analysis the genes involved in “organization of extracellular matrix,” such as LGALS3, are enriched in old TauTKO hearts compared to old wild-type hearts, suggesting the contribution of these genes to fibrosis. In conclusion, taurine depletion predisposes the heart to fibrosis, which leads to cardiac fibrosis upon aging.
Taurine is abundant in mammalian tissue, especially in excitable tissues such as the heart (about 20 µmol/g tissue). Taurine possesses various cellular actions, such as the regulation of osmolality, modulation of ion movement and calcium handling, modulation of neurotransmission, etc.1–3) The high intracellular taurine pool is maintained through both the diet and biosynthesis in the liver and adipose tissue.4) In humans, meat and seafood are important sources of dietary taurine.5) Taurine has also been used as an effective pharmaceutical agent against a wide variety of cardiovascular diseases, including chronic heart failure.1) Impetus for the pharmacological use of taurine came from studies showing that a taurine deficient diet leads to severe taurine deficiency in certain species, such as cats and foxes, and results in several tissue disorders, including dilated cardiomyopathy.6–8)
Many mechanisms are involved in taurine depletion-related cardiomyopathy.9) First, taurine depleted heart exhibits abnormal energy metabolism, including the activation of glycolysis.10) Second, taurine depletion causes a reduction in calcium handling activity, which relates to systolic and diastolic defects.11) Third, deficits in Ca2+ sensitivity of myofibril also observed in taurine-depleted heart.11,12)
Cardiac fibrosis is a major cause of cardiac dysfunction in a variety of diseases, including hypertension, cardiomyopathy and cardiomyositis.13) Cardiac fibrosis accompanying advanced aging is also an important mechanism of cardiac dysfunction.14,15) Accumulation of extracellular matrix leads to a progressive increase in cardiac stiffness and diastolic dysfunction. Although it is logical to assume that cardiac fibrosis could contribute to aging-dependent cardiac dysfunction in taurine-depleted cardiomyopathy, the possibility that taurine depletion might cause cardiac fibrosis has not been investigated.
We previously reported that the knockout of the taurine transporter (TauT) results in severe taurine depletion in the heart and results in the development of cardiomyopathy characterized by atrophy, ventricular wall thinning and induction of heart failure marker genes in mice (TauT-knockout (TauTKO) mice).16) A reduction in cardiac output has been observed in old (>9-month-old) TauTKO mice, but not in young TauTKO mice. Therefore, in the present study, the occurrence of aging dependent cardiac fibrosis was examined in TauTKO mice. Also examined was the mechanism underlying cardiac fibrosis in older TauTKO mice using transcriptome-pathway analysis.
Animal studies were carried out in accordance with an animal protocol approved by the Institutional Animal Care and Use Committee of Hyogo University of Health Sciences and the Institutional Animal Care (Approval numbers: 2009-06, 2011-07, 2013-08).
Mice were euthanized by CO2 inhalation or cervical dislocation, and all efforts were made to minimize suffering of the animals. TauTKO and littermate mice (C57BL/6 background) were obtained by breeding heterozygous male and female.16) Mice were housed in a Specific pathogen-free environment, fed standard chow (MF, Oriental Yeast, Japan), had access to water ad libitum and maintained on a 12-h light/dark cycle. Young (3 to 5-month-old) and old (17 to 24-month-old) mice were euthanized and removed for analysis.
Histological AnalysisSections from frozen tissues were cut by cryostat (Carl Zeiss, Germany). Sections were stained by Hematoxylin & Eosin method or Picro Sirius red stain (Picrosirius Red Stain Kit, Polysciences, Inc., Germany). The fibrotic area to total area was quantified using Scion Image software (Scion Corporation, U.S.A.).
RNA Isolation and Real-Time PCRTotal RNA was isolated from hearts of TauTKO and wild-type (WT) mice by using Sepazol (Nacalai Tesque, Japan). cDNA was generated from total RNA by reverse transcription using Rever Tra Ace (Toyobo, Japan) as previously described.17) Quantitative RT-PCR analyses were performed by using Applied Biosystems Step One Plus (Thermo Fisher Scientific, U.S.A.) with THUNDERBIRD SYBR qPCR Mix (Toyobo). The primers used are listed in Table 1. follows; Col1a1 Forward 5′-AAC AGT CGC TTC ACC TAC AGC AC-3′, Reverse: 5′-CGG GAG GTC TTG GTG GTT TT-3′, Col1a2 Forward: 5′-CTG AGG GCA ACA GCA GGT TC-3′, Reverse: 5′-GGC AGG CGA GAT GGC TTA TT-3′, Col3a1 Forward: 5′-TCA AGG CTG AAG GAA ACA GCA-3′, Reverse: 5′-TCT TGC TCC ATT CCC CAG TG-3′.
| Functions annotation | p-Value | Molecules |
|---|---|---|
| Muscle contraction | 1.81E-09 | ACTA2, CACNA1H, CACNA1S, CASQ1, CNN1, CTGF, FKBP1B, GJA5, GNAO1, KLK3, MYBPC2, MYH11, MYH7, MYL1, MYL9, MYOT, PTHLH, SCN4B, STBD1, TBXA2R, TMOD4, TNNT3 |
| Heart rate | 3.65E-07 | ACTG2, ADRA2C, APLN, CACNA1H, CTGF, DDAH1, FKBP1B, GJA5, GNAO1, ITGB3, KCNIP2, KLK3, MYH7, MYL1, NPPA, Nrg1, PTHLH, PTS, RNLS, SCN4B, SFRP2 |
| Transport of glycine | 3.26E-05 | SLC36A2, SLC38A2, SLC6A17, SLC6A9 |
| Invasion of cells | 3.74E-05 | ACTA2, AQP8, BCL2, CA3, CDKN2A, COL18A1, CTGF, DKK3, ENAH, ETV5, FBLN2, GDF15, GLI1, HPSE, ITGB3, ITGB4, ITGB6, KLK3, LGALS3, MMP3, PLXNB1, PTK2B, RRM2, S100A4, S100A9, SDC4, SERPINA1, SFRP1, SLIT2, TBXA2R, TIMP1, UBD, WISP1, WISP2 |
| Proliferation of cells | 4.19E-05 | ACHE, ADAMTS8, AHNAK, ALDH3A1, ALOX12, ANGPT1, APLN, BCL2, CA3, CACNA1S, CAMKK1, CD72, CDA, CDKN2A, CENPF, CGREF1, CITED1, CLDN15, CMA1, CNN1, COL18A1, COMP, CTGF, CTH, DFNA5, DHH, DKK3, DTX1, DUSP4, EPHB1, ETV5, EXTL1, FBLN2, FBXO2, FKBP1B, GCGR, GDF15, GJA5, GLI1, GNAO1, GNG2, GNMT, GPNMB, GRIN2C, HPSE, IFT122, INTU, IRF6, ITGB3, ITGB4, ITIH4, KIF1A, KIT, KLK3, LAMC3, LECT1, LGALS3, LGI1, LRRC32, MAP1B, MAS1, MLXIPL, MMP3, MYH11, MYL9, NCAM1, NGEF, NOX4, NPPA, Nrg1, NRG4, PCGF2, PHLDA1, PLXNB1, PNP, PPAP2C, Prg4, PRKAR2B, PRRX2, PTHLH, PTK2B, PTPRF, RAB17, RRM2, S100A4, S100A8, S100A9, SCN4B, SELL, SEPT6, SERPINA1, SFRP1, SFRP2, SLC1A2, SLC22A1, SLIT2, SLIT3, SP110, SPRR1A, STAM2, STK38, STXBP4, TBXA2R, TIMP1, TMEM35, TNFRSF19, Tnfsf9, TRIM67, TRO, UNC119, UPP1, VSIG4, VWF, WIF1, WISP1, WISP2, WNK2, ZSCAN18 |
| Organization of extracellular matrix | 4.08E-03 | APLP1, COL18A1, COMP, KAZALD1, LGALS3, Nepn, SMOC1 |
RNA from heart was isolated by using Sepazol-RNA super G, and cleaned by using RNeasy mini kit (Qiagen, Germany). A microarray analysis was performed on 4 groups (Old-TauTKO, Old-WT, Young-TauTKO and Young-WT (n=3 for each group)) by using SurePrint G3 Mouse Gene Expression 8x60K arrays (Agilent Technologies, U.S.A.) according to the manufacturer’s instructions. The microarray data was deposited in Gene Expression Omnibus. Data analysis was performed with GeneSpring software (version GX 12.6, Agilent Technologies). All comparisons of expression levels between the groups were performed using moderate un-paired t-tests. The genes whose levels were called more than 100 in all three samples of at least one group were subjected to following analysis. Genes were identified as differentially expressed if they showed a fold-change of at least 1.8 with a p-value lower than 0.05 between groups.
Pathway analysis was performed using Ingenuity Pathway Analysis (IPA) software (QIAGEN). The Fisher’s exact test was used to determine statistical significance for association with recorded knowledge concerning molecular networks, upstream regulators and biological functions in IPA.
StatisticsStudent’s t-test or Tukey–Kramer test (for multiple comparisons) was used to determine statistical significance between groups. Data were expressed as means±standard error (S.E.). Differences were considered statistically significant when the calculated p value was less than 0.05.
We analyzed whether cardiac fibrosis progresses in TauTKO mice during aging. Histological analysis revealed that the interstitial space increased more in the heart of old TauTKO mice than in their WT cohorts, whereas fibrosis was not observed in the heart of young TauTKO mice (Fig. 1A). Picrosirius red stained-section showed interstitial fibrosis was 14-fold greater in old TauTKO mice than in old WT mice (Fig. 1B).

(A) Representative images of hematoxylin–eosin staining (HE) and Picrosirius Red staining (PSR) are shown. Scale bars=100 µm. (B) The ratio of fibrotic area to total area was calculated (n=4). * p<0.05. (Color figure can be accessed in the online version.)
To clarify whether collagen biosynthesis is activated in old TauTKO heart, mRNAs of collagen type 1 and 3 were analyzed by real-time PCR. Significant changes in both collagen type 1 (col1a1 and col1a2) and type 3 (col3a1) were not detected between old TauTKO and old WT mice (Fig. 2), indicating that collagen transcription was not activated in old TauTKO hearts.
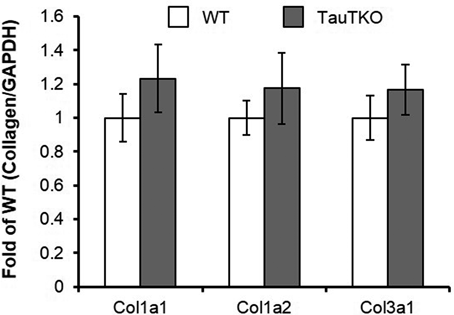
Expression levels of collagen (Col) 1a1, 1a2 and 3a1 mRNA were analyzed. Collagen mRNA level was normalized by GAPDH mRNA level. n=5.
To reveal the molecular basis of aging-associated fibrosis in TauTKO mice, microarray analysis was performed on ventricles of young and old TauTKO and WT mice (n=3) (Fig. 3A). We defined the genes that are expressed more or less than 1.8-fold with a p-value <0.05 compared to WT as differentially expressed genes. Although 590 genes were identified as significantly different between old TauTKO and old WT while 414 genes showed significant differences between young TauTKO and young WT, 240 genes exhibited overlap between the two gene sets (Fig. 3B).

(A) Volcano plot of microarray data of heart from TauTKO and WT mice at young age and old age. The log2 fold change is displayed on the x-axis and –log 10 (p-value) on the y-axis. The black dots indicate genes that are differentially expressed with a p-value >0.05. The other genes are indicated by gray dots. (B) Venn diagram of the overlap among significantly altered genes in old TauTKO compared to old WT and of young TauTKO compared to young WT hearts (>1.8-fold change). (C, D) The expression levels on genes for extracellular matrix processing (C) and fibrogenic genes (D) are shown. n=3. Data were expressed as means±S.E. *; p<0.05, **; p<0.01 between two group.
Concerning extracellular matrix processing, proline-4-hydroxylase 1 (P4ha1), matrix metalloproteinase 3 (Mmp3) and serine protease inhibitor A1 (Serpina1a, c, d and e) were induced in old TauTKO mice (Fig. 3C). Moreover, tissue inhibitor of metalloproteinase 1 (Timp1) tend to be induced in old TauTKO mice.
Importantly, pro-fibrotic genes, such as Acta2, S100a4 and Ctgf, were increased both in young and old TauTKO compared to age-matched WT hearts (Fig. 3D). In addition, the expression level of genes involved in pro-fibrogenic factors, angiotensin II, endothelin-1 and transforming growth factor (TGF)-beta family varied. In TauTKO hearts, angiotensin II- and endothelin-related genes (Ace, Agt, Agtr1a, Ece, Edn1, Ednra, Ednrb) were not changed compared to its WT cohorts (Data not shown). However, in the TGF family, Gdf15 levels were 5-fold higher in TauTKO mice than in WT mice, whereas TGF-beta1, 2 and 3 were only 1.0–1.4 fold higher in the taurine deficient heart. Furthermore, consistent with our previous report,16) cardiac failure markers, such as ANP, BNP and βMHC, were induced in both young and old TauTKO hearts.
Pathway Analyses in Transcriptome Profile of TauTKO and WT HeartsPathway analysis (IPA) using microarray data was carried out to reveal the molecular mechanism for taurine depletion-related cardiomyopathy and aging-associated cardiac fibrosis. The analysis of biological function with the gene set differentially expressed between old TauTKO and old WT mice revealed significant enrichment of genes involved in “muscle contraction,” “heart rate,” “transport of glycine,” “invasion of cells,” “proliferation of cells” and “organization of extracellular matrix” (Fig. 4A, Table 1). Furthermore, the fold changes of genes involved in “organization of extracellular matrix,” such as Aplp1, Col18a1, Comp, Kazald1, Lgals3, Nepn, Smoc1, among 4 groups are focused (Fig. 4B). Aplp1, Comp, Nepn and Lgals3 are highest in old TauTKO heart in 4 groups, while the other genes are increased both in young and old TauTKO heart compared to age-matched control.

(A) Microarray data containing genes differentially expressed between Old TauTKO and Old WT hearts (>1.8-fold) were analyzed by Ingenuity Pathway Analysis (IPA) software for functional analysis. Detailed analysis information is listed in Table 1. (B) Fold changes in genes involved in “organization of extracellular matrix” are shown. n=3, *; p<0.05, **; p<0.01 between two group.
In this study, we analyzed the potential contribution of cardiac fibrosis toward the aging-dependent decline in cardiac function of the TauTKO mouse. Enhanced deposition of collagen in the interstitial matrix was observed in old, but not young, hearts of TauTKO mice. However, mRNA expression of collagen type 1 and 3 genes were not increased in old TauTKO. Fibrosis is one of the features of aging in the heart as well as in several cardiovascular diseases, including hypertension and myocardial infarction. While elevation of collagen mRNA is reported in animal models of cardiovascular diseases, such as myocardial infarction and hypertension, collagen type 1 and 3 mRNA levels do not increase with advancing aging in the heart.15) Therefore, the mechanism of fibrosis in old TauTKO mice may be related to the pathway involved in aging-dependent fibrosis. The mechanisms underlying the process of aging-dependent cardiac fibrosis have not fully elucidated, while post-translational processes are associated with aging-dependent cardiac fibrosis.15) While glucose-mediated formation of advanced glycation end-products (AGE) is increased in the aged tissues, collagen glycation with aging may be important to collagen accumulation in the aged heart.15) Moreover, inhibition of AGE formation by AGE cross-link breaker improved cardiac function in the aged animals.15,18) Taurine possesses the preventive action against AGE formation due to high reactivity with aldehyde.19,20) It has been reported taurine treatment prevents an increase in the plasma glycated proteins in high fructose-fed rats.20) Furthermore, taurine treatment ameliorates fructose diet-induced collagen accumulation and extensive cross-link between collagen fibers in skin.21) Therefore, endogenous taurine potentially plays a preventive role in collagen cross-link by preventing AGE formation.
Moreover, microarray analysis revealed fibrosis-related gene expression was activated in TauTKO hearts. A member of the serine protease inhibitor family, Serpinal (alpha-1 antitrypsin) is elevated in only old TauTKO hearts. Serpina1 can inhibit several proteases, such as trypsin, chymotrypsin, thrombin, plasmin, cathepsin G, kallikrein, proteinase 3, renin, and pancreatic elastase.22) Importantly, Serpina1 can cause a change in collagen metabolism.23) While the role of Serpina1 in fibrosis has never been reported, other protease inhibitors, plasminogen activator inhibitor-1 (PAI-1) and tissue inhibitor of metalloproteinase-1 (TIMP-1) contribute to the reduction in collagen degradation by inhibiting matrix metalloproteinases.13) Thus, Serpinal may potentially play a key role in aging-dependent cardiac fibrosis, at least, in TauTKO mice. Additionally, the present study showed the induction in P4HA1 expression in old TauTKO hearts, which may contribute to activation of post-translational modification of collagen.
The pathway analysis based on microarray data reveal that the enrichment of genes involved in “organization of extracellular matrix” may contribute to the progression of fibrosis. Importantly, the expressions of APLP1, LGALS3 and Cartilage Oligomeric Matrix Protein (COMP) in old TauTKO hearts are more than 2-fold higher than in both old WT and young TauTKO hearts, suggesting that these genes may play important roles in the progression of aging-associated cardiac fibrosis. The role of LGALS3 (galectin-3) in promotion of fibrosis is well established. It has been reported that hepatic and lung fibrosis, which are induced by CCL4 and TGF-beta, respectively, are blocked in LGALS3 knockout mice.24,25) The expression of LGALS3 is induced in heart failure in both human patients and animal models,26) suggesting a contribution of LGALS3 toward cardiac fibrosis. Importantly, in human clinical study, circulating level of LGALS3 was associated with all-cause mortality,27) suggesting that elevated LGALS3 in old TauTKO heart may contribute to shortening lifespan.28) COMP, also known as thrombospondin-5, is a member of the thrombospondin family, and has been reported to interact with extracellular matrix proteins. It has been demonstrated that COMP catalyzes collagen fibril formation.29) Meanwhile, expressions of Col18a1, KAZALD1 and SMOC1 are higher in young and old TauTKO mice than in age-matched WT mice, indicating that endogenous taurine depletion may directly cause to high expression of these genes. We found such potential genes which can be associated with cardiac fibrosis in TauTKO mouse. However, we could not conclude whether these genes associate with cardiac fibrosis due to technical limitations. For example, while the products from these genes localize in extracellular matrix, the role for fibrosis have not been evaluated.
In conclusion, tissue taurine depletion increases the susceptibility of the heart to aging-dependent cardiac fibrosis.
We thank Messrs. S. Tanaka, S. Ueno and colleagues for their works for animal care. This work was granted from the JSPS KAKENHI Grant Numbers 22790097, 25750368.
The authors declare no conflict of interest.