2018 Volume 41 Issue 12 Pages 1866-1869
2018 Volume 41 Issue 12 Pages 1866-1869
Myeloid-derived suppressor cells (MDSCs) are immunosuppressive myeloid cells found in patients with cancer and in mouse tumor models. They suppress anti-tumor immunity, resulting in the promotion of tumor growth. The relationship between nutrition and cancer has recently been reported by several research groups. Tumor cells rely on glutaminolysis, in which glutamine is metabolized into glutamate for energy production, and hence, glutamate levels are elevated in tumor-bearing hosts. However, the mechanism of regulation of tumor progression by glutamate still remains unclear. In this study, we found that the metabotropic glutamate receptor (mGluR) 2/3 was expressed on MDSCs, and an mGluR2/3 antagonist LY341495 attenuated the immunosuppressive activity of MDSCs. Furthermore, we observed that LY341495 treatment inhibited B16-F10 melanoma growth in vivo. Taken together, our data suggest that glutamate signaling promotes tumor growth by increasing the potency of immune suppression.
Multiple studies have revealed that immune suppression in patients with cancer enhances tumor progression. Myeloid-derived suppressor cells (MDSCs) are immunosuppressive myeloid cells that accumulate in tumor-bearing hosts and promote tumor growth by inhibiting anti-tumor immunity.1,2) MDSCs reportedly suppress T cell proliferation via arginase-, inducible nitric oxide synthase-, or reactive oxygen species-dependent mechanisms.1,3,4) However, the mechanisms of the differentiation and immunosuppressive function of MDSCs are not completely understood.
High glutaminase and low glutamine synthetase activity have been observed in several types of tumor cells, and elevated glutamate levels in plasma and tumor tissues have been reported in tumor-bearing animals.5,6) Therefore, MDSCs are speculated to be exposed to high glutamate concentrations. Based on this, we hypothesized that glutamate enhances tumor progression by inducing MDSCs or enhancing their immunosuppressive function.
The metabotropic glutamate receptors, mGluR2 and mGluR3, have been shown to play crucial roles in anxiety and stress disorders,7) but their roles in immune cells have not yet been elucidated. In this study, we evaluated the expression of mGluR2 and mGluR3 in MDSCs in vitro. We found that LY341495, an mGluR2/3 antagonist, attenuates the immunosuppressive function of MDSCs in vitro. Furthermore, LY341495 inhibits B16-F10 melanoma growth. Taken together, these results suggest that LY341495 would enhance anti-tumor immunity in B16-F10 melanoma-bearing mice through attenuation of the immunosuppressive function of MDSCs. Thus, glutamate might play important roles in enhancement of the immunosuppressive function of MDSCs in tumor-bearing hosts via mGluR2/3, resulting in tumor progression.
C57BL/6J mice were purchased from Japan SLC (Hamamatsu, Japan) and used at 6–10 weeks of age. All animals were bred under specific pathogen-free conditions. All animal experimental procedures used in this study were performed in accordance with the institutional guidelines for animal experiments at the Osaka University (Approval: Douyaku 28-3-1 and 28-8-3).
Cell Preparation and in Vitro MDSC DifferentiationSpleens were dissected, passed through a 70 µm cell strainer (Corning, U.S.A.), and washed with 2% fetal bovine serum (FBS)/phosphate buffered saline (PBS) thereafter. Splenocytes were isolated, followed by red blood cell lysis using a lysis buffer (1 M NH4Cl, 0.1 M KHCO3, and 1 M ethylenediaminetetraacetic acid). Bone marrow (BM) was flushed aseptically from femurs, using 2% FBS/PBS and a syringe, and red blood cells were lysed with the lysis buffer. The isolated BM cells were cultured in the presence of 40 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF) (PeproTech) for four days; the obtained cells were used as in vitro generated MDSCs. LY341495 (Tocris Bioscience) was added daily at a concentration of 10 µM for four days during the in vitro MDSC differentiation.
Flow CytometryMononuclear cells were stained with antibodies for 20 min at 4°C. The following specific antibodies were used: CD11b-PE-Cy7, Gr-1-FITC/APC, and Ly-6G-APC from BioLegend; Ly-6C-V450 from BD Biosciences; and mGluR2- and mGluR3-rabbit polyclonal antibodies from Alomone Labs. Phycoerythrin (PE)-labeled donkey anti-rabbit immunoglobulin G (IgG) (minimal x-reactivity) antibody (BioLegend) was used as a secondary antibody for mGluR staining. Dead cells were removed by adding 7-AAD viability staining solution (eBioscience). Flow cytometry analysis was performed using a MACS Quant Analyzer (Miltenyi Biotec) and FlowJo software (TOMY Digital Biology).
Isolation of CD4+ T and CD8+ T CellsSplenocytes were incubated with anti-CD4 or anti-CD8 Microbeads (Miltenyi Biotec) to enrich CD4+ and CD8+ T cells, respectively, and positively selected using an autoMACS Pro Separator (Miltenyi Biotec). The purity of each enriched cell population was confirmed to be >90% by flow cytometry.
T Cell Suppression AssaySplenocytes (1×105), labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE; Dojindo, Kumamoto, Japan) or eFlour 670 (eBioscience), were co-cultured with various amounts of MDSCs in a 96-well plate for three days in RPMI 1640 medium (Sigma, U.S.A.) supplemented with 10% FBS, 10 mM N-(2-hydroxyethyl)piperazine-N′-2-ethanesulfonic acid (Gibco), 100 µM non-essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), 55 µM 2-mercaptoethanol (Gibco), 2 mM Glutamax (Gibco), 100 U/mL penicillin, and 0.1 mg/mL streptomycin. T cell proliferation was induced by stimulating the cells with 1 µg/mL of anti-CD28 (BioLegend) in a 96-well plate pre-coated with 1 µg/mL of anti-CD3ε (BioLegend). T cell proliferation was measured, using CFSE or eFlour670 dilution, with a MACS Quant Analyzer (Miltenyi Biotec) and FlowJo software (TOMY Digital Biology).
Tumor InoculationC57BL/6J mice were injected intradermally in the flank with 1×106 B16-F10 cells. The tumor volume was calculated according to the equation: [length×(width)2]/2. LY341495 (3 mg/kg) or dimethyl sulfoxide (DMSO) (1.76 mL/kg) was administered every alternate day from day 1 to day 9. Mice were analyzed at day 14.
Statistical AnalysisResults are expressed as the mean±standard error of the mean. Differences were analyzed by Student’s t test or two-way ANOVA, for comparison between two groups, and one-way ANOVA, for comparison among three or more groups, by using GraphPad Prism software (GraphPad Software).
The relationship between nutrition and tumor progression has been noted for many years. Altered nutrient balance likely contributes to tumor progression; however, the mechanisms connecting the two are not well understood. The levels of most amino acids, including glutamate, in tumor tissues obtained from patients with colon and stomach cancers have been reported to be significantly higher than those in the normal colon and stomach tissues of patients.5) These findings imply that amino acids could function as biomarkers of cancer and could regulate tumor progression.
Group II mGluRs (mGluR2 and mGluR3) has been reported to have a functional role in supporting the growth of U87MG human glioma cells both in vitro and in vivo.8,9) However, little attention has been paid to the functional role of mGluR2/3 in supporting the immune suppression and growth of other tumors.
Previous microarray data10) showed that, out of all glutamate receptors, mGluR3 was expressed more in MDSCs than in neutrophils. In this study, we observed that mGluR2 and mGluR3 were expressed at low levels on MDSCs in vitro (CD11b+Gr-1+ cells) (Fig. 1). The mGluR2/3 antagonist, LY341495, did not affect the proportion of CD11b+Gr-1+ cells and two murine MDSC subsets, polymorphonuclear (PMN)-MDSCs (CD11b+Gr-1highLy-6G+Ly-6C+) and monocytic (M)-MDSCs (CD11b+Gr-1midLy-6G−Ly-6C+) (Fig. 2a). In vitro MDSCs, differentiated in the presence of LY341495, could not sufficiently suppress the proliferation of T cells (Fig. 2b). We found a difference in the reactivity between CD4+ and CD8+ T cells in T cell suppression. It is suggested that MDSCs suppress the proliferation of CD4+ and CD8+ T cells by different mechanisms that are yet to be revealed. Further, we found that B16-F10 melanoma tumor growth rate was reduced by LY341495 in B16-F10 melanoma-bearing mice that had received LY341495 intraperitoneally (Fig. 3). Taken together, our data suggest that LY341495 enhances anti-tumor immunity via the inhibition of MDSCs in B16-F10 melanoma-bearing mice.
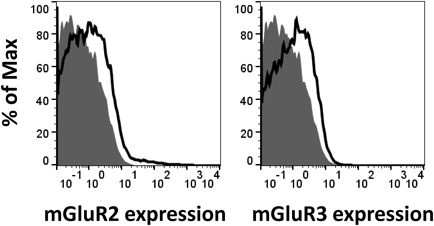
Histograms show the staining of mGluR2 and mGluR3 (black lines), and isotype controls (grey shading); data were gated on CD11b+Gr-1+ cells and are representative of two independent experiments.

(a) Markers of MDSCs, PMN-MDSCs, and M-MDSCs in MDSCs, differentiated in the presence of DMSO or LY341495 in vitro, with GM-CSF for 4 d were analyzed by flow cytometry (n=4, Student’s t test; n.s., not significant). (b) The in vitro MDSCs differentiated in the presence of LY341495 or DMSO were co-cultured with eFlour670-labeled CD4+ or CD8+ T cells, followed by stimulation with anti-CD3ε/CD28 antibodies for 3 d. The ratio indicates MDSCs to T cells. NC: negative control, PC: positive control. The proportion of proliferated T cells in each PC group was 100%, based on which the relative proliferation of each group was calculated. The values represent the means±S.D., in triplicate. Data are representative of four independent experiments (one-way ANOVA, * p<0.05, ** p<0.01).
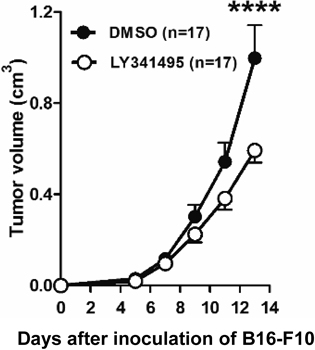
Tumor growth in LY341495- and DMSO-treated mice following implantation of B16-F10 melanoma cells. DMSO or LY341495 was administered every alternate day from day 1 to day 9 (n=17, two-way ANOVA, **** p<0.0001).
Although further research is needed, our findings suggest that glutamate regulates tumor progression via the immune system; targeting MDSCs might enhance the efficiency of immune checkpoint blockade therapy.
This work was supported in part by the SEI Group CSR Foundation, Mishima Kaiun Memorial Foundation, SENSHIN Medical Research Foundation (to M. T.), KAKENHI Grants-in-Aid for Young Scientists (B) (Grant number JP16K08268 to Y. A.) from the Japan Society for the Promotion of Science (JSPS), and the Takeda Science Foundation (Japan) (to Y. A.). This research was partially supported by the Platform Project for Supporting Drug Discovery and Life Science Research funded by the Japan Agency for Medical Research and Development (AMED) under Grant number 18am0101084j0002.
The authors declare no conflict of interest.