2018 Volume 41 Issue 9 Pages 1311-1321
2018 Volume 41 Issue 9 Pages 1311-1321
In biological systems, extracellular vesicles including exosomes have recently been revealed to play a significant role in the communication between various cells, and the number of papers on this subject has dramatically increased. In current conventional exosome studies, the standard research method is to use liquid biopsies to analyze extracts of various disease exosomes. However, exosomes are only one of many key players in natural cellular interactions. Reproducing the phenomena occurring in vivo and investigating the interactions are required in order to examine their role fully. For exosome research, an alternative to the liquid biopsy method for observing natural interactions is the co-culturing technique. It does not require an exosome extraction procedure, and while the technique has been used in many studies thus far, its application to exosome research has been limited. However, the use of co-culturing technologies is necessary to examine the essential interactions of exosomes. An overview of exosome research methodologies and co-culturing systems is thus provided here.
A wide variety of cell types are present in the human body. Individual cells do not exist in isolation but communicate with each other and can function as a collective in vivo; moreover, interactions among the collectives allows performance as a single entity. Until recently, intercellular communication was considered to be conducted through direct interactions and soluble molecules such as cell-secreted cytokines. However, in addition to these mechanisms, more recent studies have revealed that extracellular vesicles are also involved in cell–cell communication. In early reports, extracellular vesicles released from platelets and red blood cells, including exosomes, were considered to be cell waste.1,2) In 1987, Johnstone et al.3) first discovered the vesicles secreted by sheep reticulocytes and labeled them exosomes. Many cells secrete exosomes and they are also present in body fluids including blood. Countless features are unknown, but proteins and nucleic acids appear to be involved. Exosomes have lipid bilayer membranes and contain tetraspanins such as CD9 and CD63, membrane transport proteins, and adhesion molecules such as integrins. Extracellular vesicles include plasma membrane-derived microvesicles, which are slightly larger than exosomes, and apoptotic bodies released during cell apoptosis.4) Although the lack of a clear definition of terms caused some confusion among researchers for a period, in recent years, the concept of exosome has been unified, mainly by the International Society for Extracellular Vesicles. Extracellular vesicles are currently divided into three main groups: 1) exosomes with lipid bilayer membranes of 50 to 200 nm in diameter; 2) microvesicles directly secreted by cells 100 to 1000 nm in size; and 3) apoptotic bodies arising from cell apoptosis.4)
Particularly noteworthy as scientific history, from 2006 through 2007, the discovery of microRNA (miRNA) and RNA in extracellular vesicles showed the possibility of genetic information transmission between cells via exosomes.5,6) Since then, researchers worldwide have turned their attention to extracellular vesicles, and exosome studies in various fields have developed rapidly. Exosomes appear to have revolutionized the conventional concept of intercellular communication; interest by researchers has accelerated, and there are many debates on the issues involved.
1.2. Survey of ExosomesExosomes, extracellular vesicles with lipid bilayer membranes, are derived from endosomes and released outside the cell. Endocytosis produces intraluminal membrane vesicles (ILVs), many of which are contained in multivesicular bodies that fuse with the cell membrane; ILVs released extracellularly are defined as exosomes.7) Characteristics of exosomes include the containment of various substances such as tetraspanins (CD9, CD63, CD81, CD82), major histocompatibility complex (MHC), stress proteins, cytoskeleton proteins, RNA (miRNA, mRNA, noncoding RNA), DNA, mDNA, and glycolipids.4,8) These substances are not included in all exosomes and differ depending on the exosome origin and state. For example, exosomes secreted from dendritic cells, which are immunocompetent cells, have both MHC class I and II molecules on their surfaces and are involved in the communication among immune cells; these molecules are not present in exosomes of other cell origins.9) With the inclusion of such substances, exosomes are responsible for intercellular communication. More recently, exosomes have also been revealed to be a means by which substances undesirable for maintaining cellular functions can be eliminated and thus they are not solely intended for communication purposes.4)
There is a consensus that exosomes are entities containing a variety of substances, enclosed by lipid bilayer membranes, and released by cells. However, there is no complete agreement among exosome researchers on answers to more basic cell biology questions regarding “what is an exosome?” There are no exact definitions of the scientific properties that distinguish exosomes from other related vesicles, and thus even the most basic questions cannot be answered. However, we are certain that these vesicles, which have been identified only recently, can demonstrate a variety of biological activities.
1.3. Exosomes and Disease ResearchCurrent exosome research is being conducted in almost all disease areas (e.g., cancer, regenerative medicine, and disorders of the immune, nervous, endocrine, and cardiovascular systems). In addition to serum and plasma in which exosomes have been extensively studied, they have also been detected in pleural effusion, breast milk, ascites, semen, salivary gland fluid, cerebrospinal fluid, and urine. Since exosomes are carriers of functional substances, they may be beneficial to the human body or have pathological effects.
An example of a reported pathological effect is the inclusion of some beta-amyloid proteins (Aβ) associated with Alzheimer’s disease into exosomes and their extracellular secretion.10) Further, induction of bronchodilation and inflammation in chronic obstructive pulmonary disease by exosomes has been noted.11–13) There are also reports that integrins contained in exosomes are predictors of future locations of metastasis14) and that melanoma-derived exosomes increase vascular endothelium permeability and promote metastasis.15) On the other hand, examples of beneficial effects to the human body include mesenchymal stem cell (MSC)-derived exosomes showing efficacy against models of acute myocardial infarction,16) acute pulmonary failure,17) and cisplatin-induced acute renal failure.17) Additionally, it has been noted that exosomes may contribute to the elimination of Alzheimer’s disease-affiliated Aβ proteins.18) Thus, exosomes are not simply intracellular entities but are also key players widely involved in concepts of disease mechanisms.
1.4. Exosomes and Drug Delivery SystemsExosomes are endogenous carriers; thus, they are considered to be potentially safer, more efficient drug delivery systems (DDS), and their utility for this application has been reported.19,20) The pharmacokinetics of exosomes differ depending on the cell origin,21) and protein analysis of the exosome membrane surface has aided in the R&D of DDS that can target specific cells.
1.5. Basic Research on Secretion and Uptake of ExosomesKnowledge to date indicates that exosomes develop from the formation of early endosomes that have been incorporated by endocytosis. Early endosomes interact with the Golgi apparatus and endoplasmic reticulum to decompose and secrete substances. Early endosomes are invaginated by limiting membranes, and then intraluminal vesicles form. This becomes the late endosome, which is released by the cell via exocytosis when it fuses with the cell membrane.22) The series of secretions are made possible by a protein complex called Endosomal Sorting Complexes Required for Transport (ESCRT). Rab2715) and neutral-sphingomyelinase27) were reported to be involved in exosome secretion. Additionally, the intracellular calcium concentration was also identified as a controlling factor of exosome secretion.23) Cells exposed to stress were shown to increase the number of exosomes released.24) The endoplasmic reticulum is drawn into recipient cells, and it was found that the mode of uptake varies and may include processes such as membrane fusion and pinocytosis.25) A detailed molecular mechanism for exosome uptake has been proposed, based on the position of phosphatidylserine (PS) inside the cell membrane but located on the outside of the exosome membrane26,27); the T-cell immunoglobulin-and-mucin-domain-containing molecule (Tim4) was found to fuse to PS.28) Other than the above, only the integrin family14) reportedly appears to be involved in the fusion and uptake of exosomes. Apart from those studies, possible components and quantitative ratios of sugar chains and lipid bilayer membranes of exosomes have been proposed but not fully elucidated. Analysis of changes after exosome uptake into cells is difficult, and the overall detailed mechanisms of secretion and uptake remain unclear.
In cancer metastasis research, regarding the premetastatic niche hypothesis that cancer cells can spread to other organs upon metastasis of the primary tumor, exosomes are proposed to play a role in the spread. Exosomes have been reported to construct premetastatic niches in advance, allowing for easier transition of cancer cells to new environments, thereby facilitating metastasis.15) In tropism of metastatic cancer cells, the mechanism controlling directional uptake appears to be hidden in the molecules expressed on the exosome surface.
Although the mechanisms controlling exosomes are gradually being revealed, our knowledge is still fragmentary. Exosome uptake and functional expression may be regulated not by a simple on/off switch but by the input of a more sophisticated “password.” Thus, to clarify these matters, analysis of the actual behavior between cells and exosome kinetics is necessary. Where do incorporated exosomes travel? How are functional substances in exosomes developed? Is exosome uptake a selective process? These issues are not yet fully understood; to elucidate these phenomena and observe interactions, a natural cell experimental system is necessary.
In exosome research, three approaches are primarily followed (Fig. 1): 1) extract exosomes and examine their contents; 2) extract exosomes and administer them to other cells to investigate the cell response; and 3) label exosomes and analyze their release into intracellular or cellular supernatants or examine their release in a co-culture system using the same method.
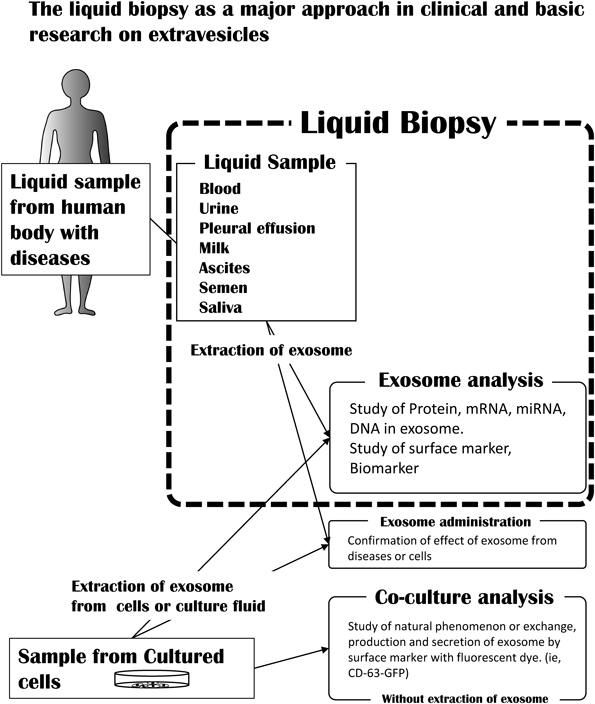
Sample fluids are blood, urine, pleural effusion, milk, ascites, semen, saliva, liquid after extraction of cultured cells, and medium of cultured cells. Three major fields of exosome study are analysis of extracted exosomes, administration of exosomes to other cells after extraction, and co-culture analysis without exosome extraction.
Until recently, approach 1), where cell-secreted exosomes are extracted and analyzed, accounted for much of the research conducted. There are numerous reports based on this approach.14,29–32) The studies focused on the analysis of functional substances contained in exosomes; the use of those substances as disease markers and drug targets for therapeutic applications is an active area of research. Of special interest is the miRNA contained in exosomes. Since the possibility of genetic information transmission between cells via exosomes was shown, miRNA in exosomes has been under intensive investigation. Successful screening for diseases using miRNA has been reported. In exosome research, many analyses have been performed on exosome inclusions that are involved in release functions, regardless of disease type, or cell function. Using information obtained with approach 1) as a supplement, research can be conducted using approach 2) to determine the effects on other cells. Regardless of whether approach 1) or 2) is followed, the extraction of exosomes is an important procedure. The results appear to be dependent on the extraction method used, and thus selection of the appropriate extraction technique is an important element in performing the research.
2.2. Exosome Extraction and AnalysisInterpretation of experimental data and confirmation of the reproducibility of exosoms research are made even more difficult due to the lack of consensus among researchers on definitions of terminology, complications in exosome detection and isolation, and obstacles in exosome purification methods. The standard extraction method for exosomes is recovery by ultracentrifugation.3,33) Other techniques include density-gradient centrifugation,5,34) polymer-based precipitation,35) immunoaffinity capture using surface markers,36) purification by flow cytometry, and size-exclusion chromatography37); however, each method has its advantages and disadvantages. It should be noted that these methods can only recover specific exosome populations due to the nature of the approach. Therefore, for research that requires comprehensive exosome recovery and for experiments aimed at extracellular vesicles including microvesicles, collection by ultracentrifugation or the density-gradient method is standard34) (Fig. 2). However, endosome-derived exosomes and plasma membrane-derived microvesicles are difficult to distinguish when ultracentrifugation is used; the possible presence of other substances also requires careful interpretation of the results.
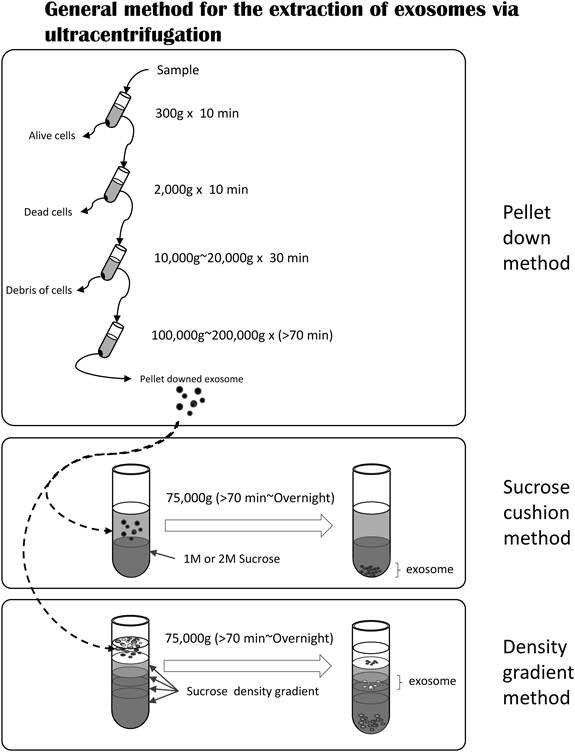
The general exosome extraction method is ultracentrifugation with or without a separating agent. The pelleting method does not use a separation agent but uses different rotation frequencies to remove cells. A separation agent is used after the pelleting method.
Apart from the exact definition by cellular origin, for convenience, extracellular vesicles that do not settle at 10000×g are often referred to as small extra vesicles or exomes.
Exosomes primarily exist in pellets after centrifugation 100000–200000×g. The use of general ultracentrifugation and sucrose density-gradient centrifugation methods is shown in a workflow diagram in Fig. 2. When the ultracentrifugation method alone is employed, the quantity of exosomes recovered is substantial, but exosome purity decreases; when the sucrose solution technique is used, exosome purity increases but the amount recovered decreases. Iodixanol instead of sucrose also can be used in a density-gradient centrifugation method.38)
After obtaining collected exosomes, particle concentration and particle size need to be verified. For these measurements, the dynamic light-scattering method is often employed. In some cases, an electron microscope can be used for verifying lipid bilayer membrane particles. The recovered and verified exosomes are subsequently concentrated and liquefied according to their intended purpose for analysis.
Particular attention is required for the experimental system and terminology used to conduct, publish, and master exosome research. A global research community in the International Society for Extracellular Vesicles has proposed “minimal information for studies of extravesicles (MISEV)” guidelines as international standards.39) Methods for documenting the experimental conditions in each publication have been proposed, and these documents need to be referenced in future exosome research papers. Current exosome research has focused mostly on their extraction and analysis, and the research techniques in many publications fall in this category.
2.3. Administering Extracted Exosomes to Other Cells or Animal Models for Observing the ResponseExperiments on administering extracted exosomes to other cells have also been performed. However, since exosomes are removed, purified, added in large quantities to other cells, and their uptake into cells analyzed in an artificial way, it is unclear if the resulting activity is truly reflective of the actual phenomena in vivo (Fig. 3A). The possibility that results due to the administration of a large quantity of purified exosomes may differ from those of the original physiological function must be noted; however, this approach is one of the established exosome research methods.18,40–45)
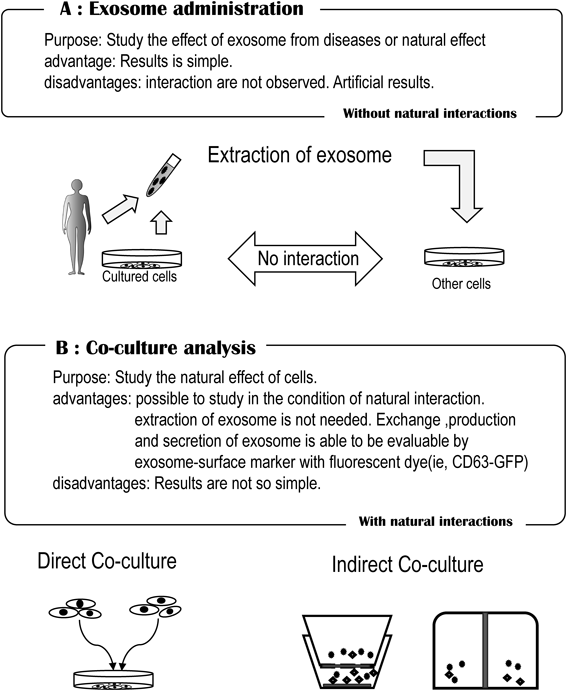
No interaction is observed in the exosome administration method. Observation of the natural interactions of cells is possible in the co-culture method.
For investigating exosome administration to other cells or animal models, the extracted exosome labeling technique is also used. Exosome imaging analysis can be performed by employing labeled fluorescent probes,21,46) and more recently, iron oxide nanoparticle probes have been used as an exosome labeling method.47,48)
2.4. Addition of Labeled Exosomes to Intracellular or Cellular Supernatants or to a Co-culture System to Examine SecretionAn alternate method to investigate intracellular dynamics and extracellular secretion without the need for exosome centrifugation is to employ fluorescent exosome surface markers such as CD63, CD81, and CD9 (Fig. 3B). The advantage of this technique is that natural interactions can be observed. However, the expression of surface markers including CD63 also depends on the type and condition of the cells. Furthermore, these proteins are not always used solely as exosome surface markers, and molecular recognition accuracy of fluorescent labels can be problematic. Although there are some limitations, for research on extracellular vesicles including exosomes, a technique using exosome labeling and a co-culture system is an important tool for studying natural cell—cell interactions. For example, by using a fusion protein of exosome-localized CD63 and green fluorescent protein (GFP), an attempt was made to visualize exosome behavior.4)
Co-culturing is a procedure whereby a plurality of different kinds of cells is grown directly or indirectly in the same environment. Co-culturing is mandatory in experimental systems that observe natural cell interactions. Since it was introduced in the 1980 s for intercellular communication research, it has been used in various fields of study. The main motivation behind employing co-culture systems is to examine interactions between natural cells and microorganisms49) and to develop novel cell engineering techniques using such interactions.50,51)
Various terms are used to refer to co-culture systems, including “coculture,” “co-culture,” “mixed culture,” “dual culture,” “co-cultivation,” and “co-incubation.” The number of times these terms were used in publications obtained through a search of the PubMed literature database (referred to below as “co-culture-related papers”) is shown in Supplementary Table 1. The total co-culture-related paper count was 101766 and the annual number of publications in this area has been increasing (Supplementary Fig. 1). Among the various terms, “coculture” or “co-culture” was used most frequently. In searching for “co-culture” or “coculture,” the number of occurrences of the two combined in documents was 25388 (40%) of 63385, with the remaining 37997 (60%) distributed between either term alone (Supplementary Fig. 2A). Since the number of documents to be searched differs greatly depending on the search term, a plurality of terms should be used for cross-sectional searches in order to uncover literature of interest. If possible, it would be desirable to unify these terms as “co-culture” or “coculture.” In the past three years, the use of “coculture” has rapidly declined (Supplementary Fig. 2B), and thus it is possible that “co-culture” will become the standard term. The various terms have made searching the literature for “co-culture” systems somewhat opaque, although co-culture systems have a long history of use in various areas of biological research.
3.2. Exosomes and Co-culture ResearchThe number of exosome research publications has been increasing dramatically (Supplementary Fig. 3A), although currently fewer (180) are found when searching for “exosome and co-culture” papers (Supplementary Fig. 3B). Considering the fact that exosomes are key players in cell interactions, analysis of natural cell interactions is an important research topic, and thus this number is expected to increase in the future.
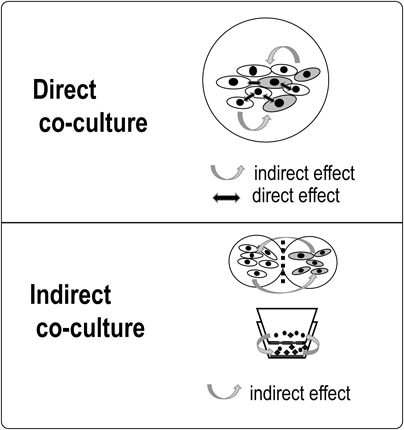
3.3. Co-culture Research MethodsExisting co-culture research methods are generally divided into two main groups depending on the state of adhesion between cells: direct co-culture; or indirect co-culture. A direct co-culture technique is used for mixing cell types together, whereby the cell types are in direct contact with each other. Although it is commonly employed, analysis of individual cells is difficult in general, and at times, changes in protein expression and the like can only be examined at the cell population level. In addition to activity through direct contact, the indirect effects of humoral factors may also interfere (Fig. 4). Thus, although it is an easy procedure to perform, interpreting the analytical methods and results is difficult. On the other hand, with indirect co-culture, cell types are placed in different environments and intercellular interactions are meditated via humoral factors. To observe intercellular interactions, culture chambers, filters, gels, etc. are employed to separate cell types that are not in direct physical contact with each other; by employing indirect humoral factors, the activity between the cell types can be clarified. Indirect co-culture systems are mainly divided into three types: 1) separation by filters; 2) separation by a semisolid such as a gel with humoral factors exchanged via the gel; and 3) formation of separate cell colonies or layers by culturing, although complete isolation of cell types is not possible (Fig. 5). Another method of exchanging conditioned media52) is also employed in microbiological research.
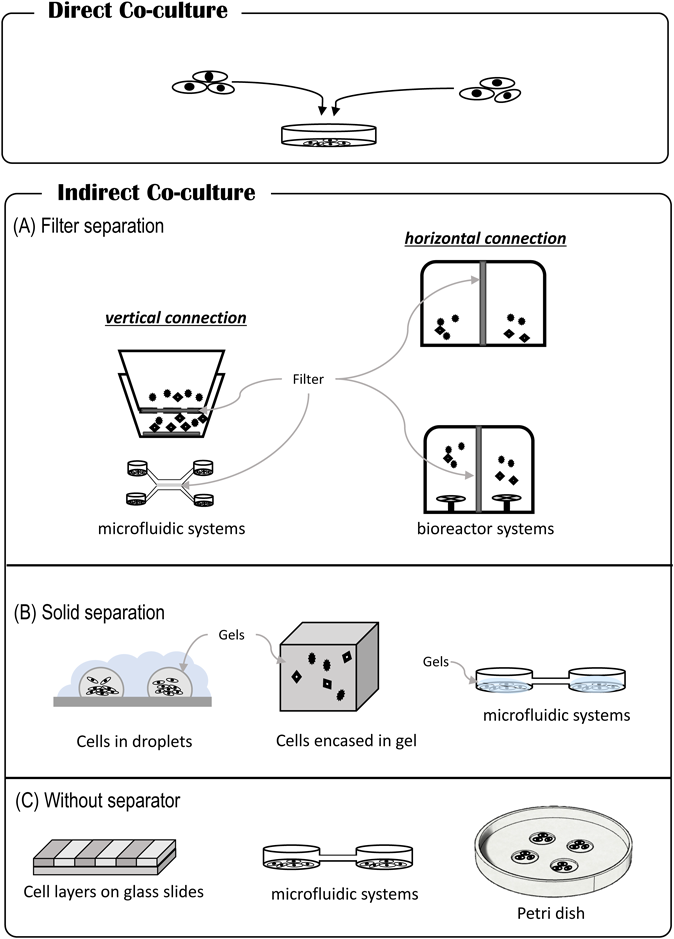
(A) Filter separation: for vertical connection, Transwell systems,64,65) microfluidic systems,51) bioreactor systems,49,66–68) and horizontal connection (this review, interactive co-culture plate, Ginreilab Inc., Japan). (B) Solid separation: cells in droplets,69–71) microfluidic system.60,72) (C) Without separator: cell layers on glass slides,73) microfluidic systems,43,74,75) Petri dish system.76) (Color figure can be accessed in the online version.)
Standard methods for filter separation of cells include methods using a Boyden chamber53–55) or Transwell co-culture plate (a modified Boyden chamber). The Boyden chamber was developed by Stephan Boyden in 1962 and is a device where the upper culture container nestles inside a lower culture container. The bottom of the upper chamber is used as the filter, and co-culturing occurs on the filter. The Transwell co-culture plate is often also called a cell culture insert. Initially, the device was employed to assess cell invasion and migration, but it is now used as a standardized method for many indirect co-culture procedures. However, it is difficult to obtain high-quality images of cells in the upper culture chamber due to the short focal length of the microscope. Another disadvantage is that the composition material of the bottom of the upper chamber is different from that of the lower chamber. Cell behavior is often affected by material surfaces, and the influence of the filter material on the bottom of the upper chamber on behavior cannot be overlooked. Moreover, since it is necessary for the filters to be integrated into the culture containers, the types of filters that can be used become limited.
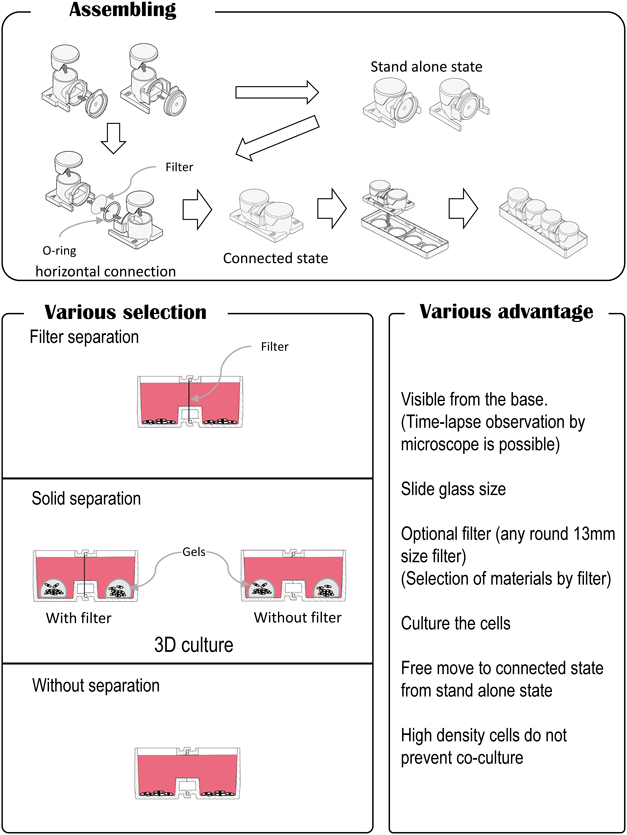
Surprisingly, almost no filter separation containers with horizontal connections have been developed. Recently, the use of horizontally connected containers for bacterial research has been reported,52) but the device was not intended for observation purposes. After we reported that the anticancer drug gemcitabine induces secretion of proteins from pancreatic cancer cell lines,56) we thought that the observation of both co-culture plates would be necessary to study the cell–cell interactions between gemcitabine-treated cancer cells and untreated ones. We then developed a novel cell-culture chamber with a transverse connection for use as a co-culture container. We refer to it as the “interactive co-culture plate (ICCP),” and the configuration allows for clearer observations with a microscope. The culture vessel is designed so that any commercially available filter can be placed between the containers. Each container can either stand alone or be connected to each other at any time. There are two ways of employing the culture vessel: 1) by initially culturing the cell types separately, then co-culturing by connecting the two containers; or 2) by joining the two containers from the beginning and using the height of the culture media to control the amount of liquid that transverses the two containers, which also controls the extent of co-culturing. The assembly of these containers is straightforward, and researchers can select any two methods of their choice. Further, since the bottom surface of the container can be examined with a microscope, individual cells can be observed simultaneously. When 3D culturing is conducted using gels or other materials, co-culturing with 3D cultures is also possible. In the Transwell type of culture vessels, the containers are fitted vertically, and cells in both the upper and lower chambers cannot be observed simultaneously with a microscope. However, since the new ICCP containers are connected horizontally, both cell types can be observed at the same time. The ICCP features and comparison with other co-culture vessels are shown in Figs. 6 and 7, respectively.
(Color figure can be accessed in the online version.)
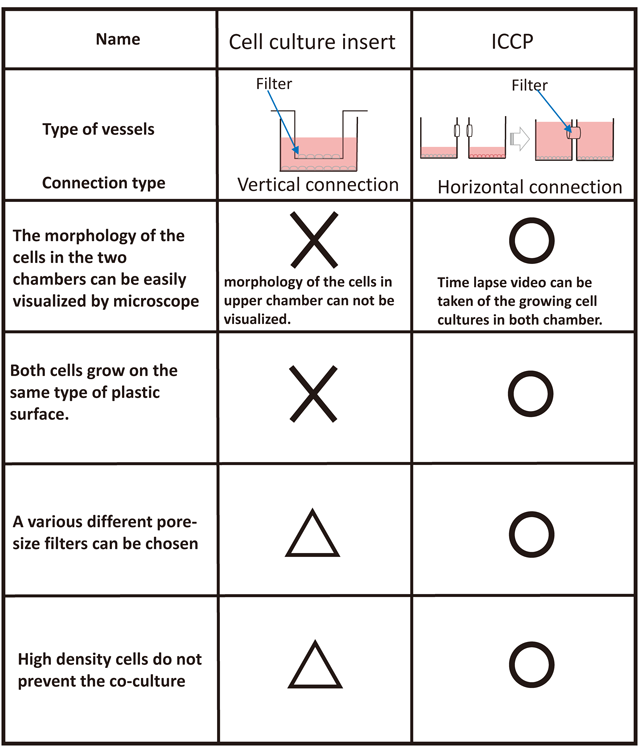
Co-culture plates with the horizontal-type connection have many advantages: researchers can observe both cell culture conditions simultaneously; time-lapse videorecordings can be made of growing cell cultures in both chambers; cells are seeded on the same type of plastic surface; pore filters of various sizes can be used; and high cell density does not interfere with co-culturing.
Regardless of the type used, co-culture systems are effective for research in which observation of intercellular interactions is necessary or cell engineering technologies that are based on cell interactions are assessed. Co-culture systems are able to provide a variety of techniques for cell engineering and they are considered particularly effective for drug discovery.51) Although high-throughput systems are required for drug discovery, constructing such systems in animal models is difficult in terms of cost and efficiency. Thus, developing a co-culture system with high-throughput capabilities is desirable in the future.
Several studies performed using direct co-culturing methods have been reported. For example, after co-culturing, exosomes derived from MSCs were taken up by breast cancer cells. Further, it was revealed that cancer cells also acquired MSC-derived cellular capacity and MSC-derived CD90.57) Fluorescent-labeling experiments that confirmed exosome exchange and migration were also reported.57–59)
In studies using indirect co-culture techniques, Transwell-type co-culture plates are important research tools. For example, a co-culture experiment on rat diabetes model cardiomyocytes and mouse endocardial cells revealed that vascular hyperplasia was suppressed and that exosomes and miR-320 contained in exosomes were involved.59) There were also reports showing that virus-derived miRNAs were transmitted to other cells via exosomes.60,61) Additionally, cells cultured above and below the filter of the Transwell co-culture plate and not in direct contact were found to change their properties through exosome activity.61,62)
Transwell co-culture plates have been reported as an in vitro blood–brain barrier (BBB) model.21) Co-culturing of cerebrovascular endothelial cells, pericytes, and astrocytes allowed for a model that simulates the BBB. These are exemplary uses of the co-culture plates.
4.2. Exosome Uptake Experiments Using the ICCP SystemWe evaluated extracellular exosome uptake using a horizontally connected co-culture system that allowed for the simultaneous observation of both culture vessels. To the gene for CD63 (one of the exosome surface markers), a gene that induced the GFP gene was introduced into cells, with resulting expression of a GFP–CD63 fusion protein and the establishment of CD63g–PANC1 cells. By selecting cells expressing red nuclei and their subsequent culture in the ICCP system, exosomes that emit GFP were detected in red-nucleated cells. When the CD63g–PANC1 cell line was cultured in one of the containers, the GFP observed was from both intracellularly produced exosomes and exosomes from neighboring cells, and thus exosome secretion and uptake could not be distinguished. However, culturing cells with GFP-labeled exosomes in only one container and detecting the presence of GFP in cells in the adjoining container allowed the confirmation of GFP passing through the filter. This means that the use of our system makes it possible to identify the origin of exosomes (Fig. 8).
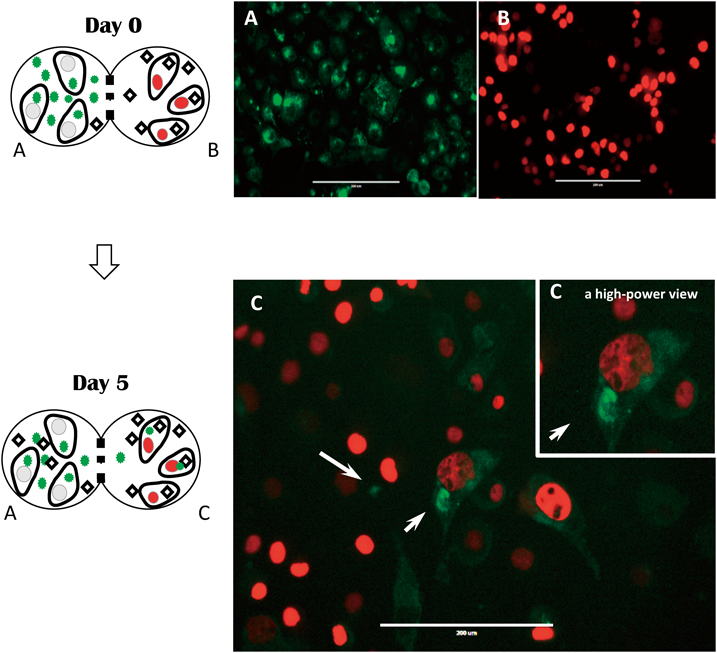
Green fluorescence was present (white arrows). Scale bar=200 nm.
We reviewed exosomes from the perspective of exosome research methods. Research is being expanded in various areas of study, including cancer, immune disorders, neurological disorders, and regenerative medicine, and currently the involvement of exosomes is becoming apparent in diseases for which the causes and mechanisms are still unknown. Although it should be noted that the exosome extraction method is not yet standardized, research on exosomes and diseases appears to indicate a relationship between disease and exosome inclusion into cells. However, knowledge is lacking on the physiological functions of exosomes and basic mechanisms of secretion and uptake. To conduct fundamental research, we believe that co-culture systems and studies on exosomes are important. By carrying out novel research using our co-culture system, we hope to elucidate the password related to exosome secretion and uptake in the future.
The authors would like to thank those who provided financial support for this project. This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number (JP23234567, JP23591016) to TS and the Kanazawa Medical University Research Project (RP2017-09) through the Private University Research Branding Project of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and in part by a Grant from MEXT (to TS). Support was also provided through Grants for Promoted Research from Kanazawa Medical University (S2013-8, S2014-7).
TS received a research Grant from Kanazawa Medical University and holds a patent (JP Patent 11201600072S) licensed to Kanazawa Medical University. TS is also President of Ginreilab Inc. SY has no conflict of interest to report.
The online version of this article contains supplementary materials.