2018 Volume 41 Issue 9 Pages 1480-1484
2018 Volume 41 Issue 9 Pages 1480-1484
Natural killer (NK) cells play an important role in tumor immunity and infection control. The natural cytotoxicity receptors (NCRs) NKp46, NKp44 and NKp30 are involved in the control of the activation of NK cells. Few reports have investigated the ligands of NCRs. We previously reported the NCRs binding affinity to heparin and glycosaminoglycans. We also showed that multimeric sialyl Lewis X-expressing transferrin, secreted by human hepatoma HepG2 cells, binds to NKp46 and NKp44, but not to NKp30. In this study, we investigated the binding between NCRs and glycolipids. The possible binding of glycolipids to NCRs was screened by microarray, using the recombinant extracellular domain of NKp46, NKp44 and NKp30 tagged with 6×His (rNKp46, rNKp44 and rNKp30). We found that rNKp44 binds to Globo-A. However, we did not detect the interaction between rNKp46 or rNKp30 and any of the glycolipids investigated. Direct binding assays supported the results of the microarray screening. Therefore, we concluded that Globo-A is a novel ligand for NKp44 but not NKp46 and NKp30, and showed differences in the ligand selectivity of NCRs.
Natural killer (NK) cells play an important role in natural immunity: they have cytotoxic activity against tumor cells and virus-infected cells, and produce cytokines for immune regulation. The natural cytotoxicity receptors (NCRs) NKp46 (also called NCR1 or CD335), NKp44 (NCR2 or CD336), and NKp30 (NCR3 or CD337) are expressed on NK cells and play an important role in recognizing target cells exhibiting little to no expression of major histocompatibility complex (MHC). NCRs have immunoglobulin (Ig)-like domains on their extracellular part and therefore belong to the Ig superfamily. As NCRs are devoid of immunoreceptor tyrosine-based activating motifs (ITAM), they transmit activation signals via associated adaptor molecules containing ITAM.1–5) NKp46 and NKp30 are expressed on both resting and activated NK cells.6–8) On the other hand, NKp44 is induced in NK cells activated by stimulation with interleukin (IL)-2.9)
The Ig-conjugated membrane-proximal domain of NKp46 (NKp46D2) and NKp44 bind to hemagglutinin of the influenza virus and hemagglutinin-neuraminidase of the Sendai virus. α2,6-Sialylation of the sugar chain on NKp46D2 and NKp44 is required for binding with these ligands. However, this modification is not necessary for their binding to tumor cells.10–12) NKp44 can also bind to Mycobacterium bovis bacillus Calmette Guerin,13) and envelope glycoproteins of West Nile and Dengue viruses.14) Microarray and surface plasmon resonance analysis with heparin oligosaccharides have revealed that NKp44 binds to highly charged and IdoA2SGlcNS6S heparin oligosaccharides, while the binding pattern of NKp46 and NKp30 to heparin oligosaccharides is different.15) Specifically, the heparin epitopes recognized by NKp46 and NKp30 are overlapping and different from those recognized by NKp44.16)
We have previously reported that K562 cells overexpressing the fucosyltransferase (FUT)-3 gene (K362/FUT3) are more susceptible to lysis by IL-2-activated KHYG-1 cells than untreated K562 cells. K362/FUT3 cells express high levels of the sialyl Lewis X (sLeX) antigen, NeuAcα2,3Galβ1,4(Fucα1,3)GlcNAc-R.17) We have also reported that the recombinant extracellular domains of 6×His-conjugated NKp46, NKp44 (rNKp46, rNKp44) can bind to multimeric sialyl Lewis X-expressing transferrin secreted by human hepatoma HepG2 cells (HepTF), while 6×His-conjugated NKp30 (rNKp30) cannot.18,19) In the same studies, we showed that basic amino acids in NKp46 and NKp44 are essential for the binding to glycan ligands such as heparin and HepTF. Additionally, we compared the binding affinity of NCRs to its ligands and showed that the Kd values of rNKp44 to heparin and multimeric sialyl Lewis X-expressing transferrin were lower than those of other NCRs, suggesting that NKp44 has high affinity for these ligands.
The elucidation of the binding of NCRs to carbohydrate ligands is important to fully understand the target recognition mechanism of NK cells. However, the knowledge of the binding of NK cells to glycolipids is poor. In this report, we screened glycolipid ligands for their binding to the recombinant NCRs using microarray, and confirmed the data obtained using direct binding assays.
The extracellular region of NKp46 (coding for amino acids [AA] 22–258), NKp44 (coding for AA 22–192) and NKp30 (coding for AA 1–135) was amplified from cDNA in human NK-derived KHYG cells (Japanese Collection of Research Bioresources Cell Bank: JCRB, Tokyo, Japan). rNKp46, rNKp44 and rNKp30 overexpressing plasmids were prepared using the pET-22b vector (TaKaRa Bio., Otsu, Japan) according to the manufacture’s instruction. The recombinant plasmids were transformed into Rosetta™ (DE3) Competent Cells (TaKaRa). rNCRs were prepared as previously described.18,19) After refolding, rNCRs were separated on 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with or without pretreatment in 1% 2-mercaptoethanol (2-ME) at 95°C for 3 min, and then stained with Coomassie Brilliant Blue. Protein concentrations were determined using the Advanced Protein Assay Reagent (Bio-Rad, Hercules, CA, U.S.A.), with bovine serum albumin (BSA) as standard.
Glycolipid MicroarrayThe binding between NCRs and various glycolipids was examined using a glycolipid array (Sumitomo Bakelite Co., Ltd., Tokyo, Japan) according to the manufacturer’s instructions. The glass slide used was coated with glycolipids (GM3, GD3, GT3, GM2, GD2, GT2, GM1a, GM1b, GD1a, GD1b, GT1a, GT1c, asialo GM1, fucosyl GM1, asialo GM2, Gb3, Gb4, Gb5, Globo-H, Globo-A, Globo-B, SSEA4-tetraose, SSEA4-hexaose and Forssman antigen). Experimental concentrations of NCRs were determined with reference to Kd values for binding to heparin as previously reported.18,19) rNKp46 (600 nM), rNKp44 (20 nM) and rNKp30 (250 nM) were added to the slide on which the glycolipids were immobilized and incubated overnight at 20–25°C. After washing three times with phosphate-buffered saline (PBS), a mouse anti-6×His monoclonal antibody (9F2; Wako Pure Chemical Industries, Ltd., Osaka, Japan) was added and incubated for 1 h at 20–25°C. After washing three times with PBS, a goat anti-mouse Cy3®, IgG H&L antibody (Abcam, Cambridge, U.K.) was added and incubated for 1 h at 20–25°C. After washing three times with PBS, the prepared slide was sent to Sumitomo Bakelite Co., Ltd. Fluorescence was measured at a wavelength of 570 nm using a model ScanArray Lite (Parkin Elmer Inc., Waltham, MA, U.S.A.). The fluorescence signals were analyzed using ProteinArray Analysis Software for ScanArray Express (ver. 4.0).
Direct Binding AssayFirst, 100 µL of rNKp44 (0–3 mM) in PBS was added to Globo-A- or BSA-coated 96-well plates (Sumitomo Bakelite) and incubated for 2 h at 20–25°C. After washing three times with Tris-buffered saline with Tween 20 (TBS-T; 20 mM Tris–HCl buffer (pH 7.4) containing 20 mM CaCl2, 0.1% BSA, 150 mM NaCl, and 0.3% Tween 20), the plate was further incubated with 100 µL of 100 mU of peroxidase-conjugated anti 6×His antibody (Wako) in TBS-T for 1 h at 20–25°C. After washing three times with TBS-T, the plates were incubated with 100 µL of tetramethylbenzidine solution (BioFX Lab., Owings Mills, MD, U.S.A.) for 10 min at 20–25°C. After the addition of 100 µL of 1 M H2SO4, absorbance was read at 450 nm using a model DTX800 plate reader (Beckman Coulter, Fullerton, CA, U.S.A.).
To investigate the binding of NCRs to glycolipids, we expressed the recombinant extracellular region of NKp46, NKp44 and NKp30 conjugated to 6×His using an Escherichia coli expression system. We separated the protein products obtained by SDS-PAGE and observed bands of about 32, 24, and 20 kDa for rNKp46, rNKp44, and rNKp30, respectively (Fig. 1).
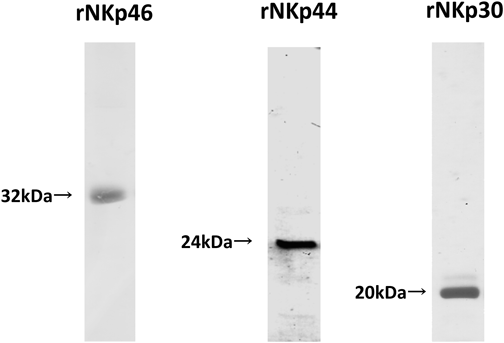
rNKp46, rNKp44 and rNKp30 purified on Ni-sepharose and refolded were separated by SDS-PAGE on 12% gels upon 1% 2-ME pretreatment.
We next examined the binding properties of NCRs to various glycolipids (Fig. 2A) using a glycolipid array, and found that rNKp44 can bind to Globo-A (Fig. 2B). The other glycolipids did not bind to rNKp44. rNKp46 (600 nM) and rNKp30 (250 nM) did not bind to the glycolipids investigated in this survey (Figs. 2C, D).
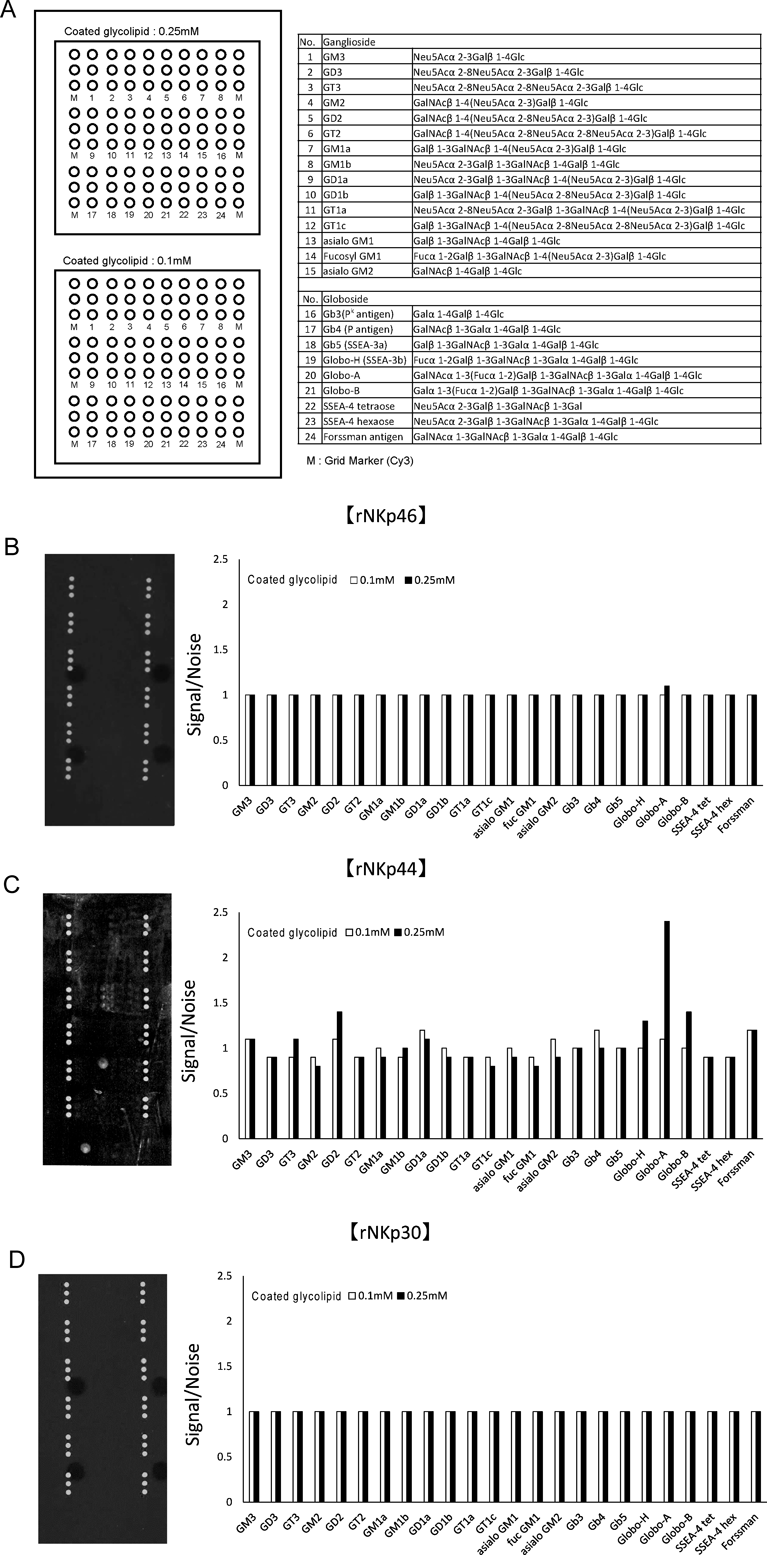
(A) Glycolipids (GM3, GD3, GT3, GM2, GD2, GT2, GM1a, GM1b, GD1a, GD1b, GT1a, GT1c, asialo GM1, fucosyl GM1, asialo GM2, Gb3, Gb4, Gb5, Globo-H, Globo-A, Globo-B, SSEA4-tetraose, SSEA4-hexaose and Forssman antigen) were coated on a glass slide at a concentration of 0.1 or 0.25 mM. (B) rNKp46 (600 nM), (C) rNKp44 (20 nM) and (D) rNKp30 (250 nM) were added to the slide and incubated overnight at 20–25°C; their binding was determined using an anti-His antibody and a Cy3-conjugated secondary antibody. Fluorescence was measured at 570 nm. The results are expressed as the ratio of signal to noise.
We next confirmed the microarray results in direct binding assays and confirmed the direct binding of rNKp44 to Globo-A. Specifically, the binding of rNKp44 to Globo-A was determined using a peroxidase (POD)-conjugated anti-6×His antibody. When 0 to 3000 nM of rNKp44 were added to the Globo-A coated plate, we observed a dose-dependent binding (Fig. 3).
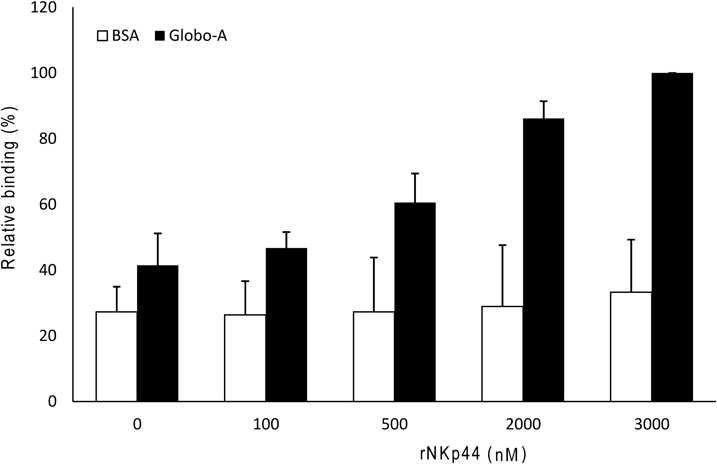
rNKp44 (0 to 3000 nM) was added to plates coated with Globo-A or BSA for at 20–25°C and binding was determined using a POD-conjugated anti-6×His antibody. The relative absorbance at a wavelength of 450 nm was normalized to the absorbance of the samples relative to the binding of 3000 nM rNKp44 to Globo-A. The results were obtained in three independent experiments and are expressed as the mean±S.D.
In this report, we screened glycolipid ligands for NKp46, NKp44, and NKp30. For glycolipid arrays, experimental concentrations of rNCRs were determined with reference to the Kd value for binding with heparin. Although it might not necessarily be the optimal concentration for binding of glycolipids to rNCRs, it was confirmed that at least Globo-A binds to rNKp44. Microarray screening showed that rNKp44, unlike rNKp46 and rNKp30, binds Globo-A, and direct binding assays confirmed this result. However, the binding affinity of rNKp44 to Globo-A was very weak and it was not possible to calculate the Kd for this interaction. Therefore, the Kd value of NKp44 and Globo-A is probably higher than that for the binding between NKp44 and other sugar chain ligands.19) In some cases, under physiological conditions, it is assumed that a large amount of ligand is present or receptors are assembled on NK cells, and strong binding may be observed.
Globo-A (GalNAcα1,3(Fucα1,2)Galβ1,3GalNAcβ1,3Galα1,4Galβ1,4Galβ1-Cer) is synthesized by 3-α-N-acetylgalactosaminyltransferase by addition of GalNAc to end of Globo-H (Fucα1,2Galβ1,3GalNAcβ1,3Galα1,4Galβ1,4Galβ1-Cer). Globo-A is known to be present in the kidney.20) Additionally, trace amounts of Globo-A are present in red blood cells.21,22) The terminal structure of Globo-A is similar to that of the blood group A antigen (GalNAcα1,3(Fucα1,2)Galβ1). In the glycolipid array results, no binding of rNKp44 to Globo-H was shown. This result suggests that the terminal part of NKp44 may be important for its binding to Globo-A. The blood group A antigen is expressed in various tissues. The activation of the expression of NKp44 in NK cells only may be related to the need of avoiding unnecessary autoimmune reactions. In previous reports, the blood group A antigen has been suggested to be involved in the progression and prognosis of several cancers. In the case of small cell lung cancer and breast cancer,23) it has been reported that disappearance of the antigen accelerates cancer progression and worsens the prognosis. Future research may be able to explain these facts by investigating the binding of NKp44 to blood group A antigen.
In this paper, we clarified that Globo-A binds to rNKp44. However, further studies are required to investigate the binding between NKp44 on NK cells and Globo-A and the effect on NK cell function. Additionally, further studies are also necessary to understand the biological significance of our results and verify whether this interaction is involved in NKp44-mediated NK cell cytotoxic activity.
This research was partially supported by the “Toho University Grant-in-Aid for Joint Research.” This work was supported by JSPS KAKENHI Grant-in-Aid for Scientific Research(C), Grant Number JP16K08945.
The authors declare no conflict of interest.