2019 Volume 42 Issue 3 Pages 481-488
2019 Volume 42 Issue 3 Pages 481-488
Lysine-specific demethylase 1 (LSD1/KDM1A) is a histone demethylase and specifically catalyzes the demethylation of mono- and di-methylated histone H3 lysine 4 (H3K4). The LSD1-mediated demethylation of H3K4 promotes the assembly of the c-Myc-induced transcription initiation complex. Although LSD1 and c-Myc are both strongly expressed in human cancers, the mechanisms by which their activities are coordinated remain unclear. We herein demonstrated that LSD1 is a direct target gene of c-Myc. The knockdown of c-Myc decreased the expression of LSD1 in several cancer cell lines. We identified two non-canonical E-boxes in the proximal promoter region of the LSD1 gene. A chromatin immunoprecipitation assay showed that c-Myc bound to these E-boxes in the LSD1 promoter. Importantly, LSD1 mRNA expression correlated with c-Myc expression in human acute myeloid leukemia (AML), glioblastoma, stomach adenocarcinoma, and prostate adenocarcinoma. The present results suggest that LSD1 is induced by c-Myc and forms a positive feedback mechanism in transcription reactions by c-Myc.
Lysine-specific demethylase 1 (LSD1/KDM1A) is a histone demethylase and member of the FAD-dependent amine oxidase family.1,2) LSD1 acts as a co-repressor by demethylating mono-methylated or di-methylated lysine 4 of histone H3.3) LSD1 also removes repressive histone marks by the demethylation of lysine 9 of histone H3, thereby promoting transcriptional activation.4) Thus, LSD1 epigenetically regulates the activation or repression of gene transcription in different cellular contexts. As a result, LSD1 is essential for stem cell function and animal development. In addition, it has been demonstrated to play an important role in cancer development.5) The overexpression of LSD1 has been reported in several types of cancers and is associated with a poor prognosis.6) Nevertheless, the regulatory mechanisms underlying LSD1 gene transcription, in normal and oncogenic contexts, have not yet been elucidated.5)
c-Myc, a member of the MYC gene family, is involved in regulating various biological activities.7) c-Myc is a basic helix-loop-helix leucine zipper (bHLH-LZ) transcription factor and dimerizes with myc-associated factor X (Max), and the c-Myc-Max heterodimer has been shown to bind to the canonical element E-box (CAC GTG and CAT GTG) or non-canonical E-box (such as CAT GCG, CAC GCG, CAC GAG and CAA CGT G) in order to activate targeted genes.8–10) In contrast, c-Myc represses various genes through its interaction with Myc-interacting Zn finger protein-1 (Miz-1). c-Myc regulates the transcription of thousands of genes required for a range of cellular processes, including proliferation, differentiation, metabolism, and stemness.11) The deregulated expression of c-Myc proteins has been implicated in the development of a wide range of cancers.12,13) Hence, the deregulated activity of c-Myc is a frequent molecular event and the inhibition of c-Myc has potential as a therapeutic strategy for cancer.14) However, the development of biologically effective inhibitors that target c-Myc is expected to be very difficult due to the absence of deep surface-binding pockets.15,16) Therefore, strategies have been extensively developed to inhibit c-Myc expression (e.g. bromodomain and extraterminal motif (BET) inhibitor), interrupt c-Myc-Max dimerization, inhibit c-Myc-Max DNA binding, and then lead to interfere with key c-Myc target genes.7)
c-Myc interacts with several histone modifiers to regulate transcription.17) Amente et al. showed that c-Myc binds to and facilitates the recruitment of LSD1 to the E-box chromatin.18) They also demonstrated that LSD1 is a cofactor of the repressive function of N-Myc (c-Myc paralog).19) Although LSD1 and c-Myc are both strongly expressed in human cancers, the mechanisms by which their activities are coordinated remain unclear. We herein demonstrated that LSD1 is a direct target gene of c-Myc. We identified two non-canonical E-boxes in the proximal promoter region of the LSD1 gene. A chromatin immunoprecipitation (CHIP) assay showed that c-Myc bound to these E-boxes in the LSD1 promoter. Importantly, LSD1 mRNA expression correlated with c-Myc expression in human acute myeloid leukemia (AML), glioblastoma, stomach adenocarcinoma, and prostate adenocarcinoma. The present results suggest that LSD1 is induced by c-Myc and forms a positive feedback mechanism in transcription reactions by c-Myc.
PC-3, HepG2, MDA-MB-231, and COS7 cells were cultured in Dulbecco’s modified Eagle’s medium (Sigma, St. Louis, MO, U.S.A.) containing 10% fetal bovine serum (Sigma) and penicillin/streptomycin.
The LSD1 promoter Luc (−830 to +113) and its deletion mutants were constructed by a PCR-based approach and cloned into pGL4.10 (Promega, Madison, WI, U.S.A.). To obtain 6xMyc responsive element (MycRE)-Luc, six tandem repeats of the E-box (CAC GTG) were ligated with pGL4.27 (Promega). pcDNA3/FLAG-Strep-c-Myc and its deletion mutant were PCR amplified using mRNA derived from MCF7 cells. All constructs were verified by sequencing.
Regarding DNA transfection, plasmids were transiently transfected with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, U.S.A.). In short interfering RNA (siRNA) transfection, siRNAs were transfected using Lipofectamine RNAiMAX reagent (Invitrogen). The human c-Myc siRNA (L-003282-00) was purchased from Dharmacon (Lafayette, CO, U.S.A.). Stealth RNAi™ siRNA Negative Control Med GC Duplex was obtained from Invitrogen.
Luciferase AssayCells were transfected with the luciferase reporter plasmid, expression plasmids, β-galactosidase (β-gal) expression plasmid, and empty vector. The total amount of transfected DNA was the same in each experiment. After 24 h of transfection, luciferase activity in cell lysates was measured using a Wallac 1420 ARVO MX (PerkinElmer, Inc., Waltham, MA, U.S.A.). Luciferase activity was normalized to β-gal activity.20)
RNA Extraction, Reverse Transcription, and Quantitative PCR (qPCR)Total RNA was extracted as described previously.21) First-strand cDNAs were synthesized with the PrimeScript first-strand cDNA synthesis kit (TaKaRa Bio Inc., Shiga, Japan) as previously described.22) q-PCR was performed as previously described.23) The following primer sequences were used: human LSD1, 5′-GGG GCT CTT ATT CCT ATG TT-3′ (forward) and 5′-CAA AGA AGA GTC GTG GAA TC-3′ (reverse); human MYC, 5′-TGT TGG TGC ACA AAA AGA CA-3′ (forward) and 5′-CAC GCC AAA CAA ATC TCC TA-3′ (reverse)24); human Telomerase reverse transcriptase (TERT), 5′-CGG AAG AGT GTC TGG AGC AA-3′ (forward) and 5′-GGA TGA AGC GGA GTC TGG A-3′ (reverse)25); human ACTB, 5′-TGG CAC CCA GCA CAA TGA A-3′ (forward) and 5′-CTA AGT CAT AGT CCG CCT AGA AGC A-3′ (reverse).26) The specificities of the detected signals were confirmed by a dissociation curve, which consisted of a single peak. Values were normalized by β-actin.
Immunochemical Methods and AntibodiesImmunoblotting was performed as previously described.27) The following commercially available antibodies were used: anti-LSD1 (#2184; Cell Signaling Technology, Beverly, MA, U.S.A.), anti-c-Myc (#13987; Cell Signaling Technology), anti-c-Myc (AF3696, R&D Systems, Minneapolis, MN, U.S.A.), and anti-β-actin (2F3) (Wako, Osaka, Japan).
ChIP AssayThe ChIP assay was performed as previously described.28) Purified DNA was analyzed by qPCR. The following primer sequences were used: human LSD1 promoter (MycRE), 5′-TAC ACG TTC TTT GCT GCG GT-3′ (forward) and 5′-ACA AAA AGG GTC GGA GAC ACC-3′ (reverse); human CCNA2 promoter (MycRE), 5′-CCA AAG AAT AGT CGT AGC CGC CG-3′ (forward) and 5′-TCG AGC TGG GTG AGC GAG C-3′ (reverse)29); human CDKN1A promoter (MycRE), 5′-TGA GCT GCG CCA GCT GAG GT-3′ (forward) and 5′-TGC CTC GGT GCC TCG GCG AA-3′ (reverse)29); human HPRT1 first intron, 5′-TGT TTG GGC TAT TTA CTA GTT G-3′ (forward) and 5′-ATA AAA TGA CTT AAG CCC AGA G-3′ (reverse).30)
RNA Expression Analysis by cBioPortalThe database cBioPortal for cancer genomics (http://www.cbioportal.org; version 1.17.1) was utilized in October 2018 to investigate the mRNA expression of LSD1 and c-Myc in various human tumor cells.31–34) The cBioPortal for cancer genomics is an open-access resource for the interactive exploration of multidimensional cancer genomic data sets.35,36)
Statistical TestsThe statistical significance of differences between two groups was evaluated using two-tailed Student’s test. For multi-group analyses, significance was assessed using one-way ANOVA with post hoc Tukey–Kramer honestly significant difference (HSD) test.
To establish whether c-Myc contributes to LSD1 regulation, we initially investigated the effects of targeting c-Myc in PC-3 human prostate cancer cells. c-Myc is present on chromosome 8q24, and PC-3 cells have the overrepresentation of 8q24.37) As shown in Fig. 1A, LSD1 and the c-Myc target TERT were down-regulated by the knockdown of c-Myc in PC-3 cells. We also found that c-Myc siRNA decreased LSD1 protein expression levels in PC-3 cells (Fig. 1B). We then investigated whether the down-regulated expression of LSD1 by the knockdown of c-Myc also occurs in other cancer cell lines without the amplification of 8q24. In the hepatoma cell line HepG2 and breast cancer cell line MDA-MB-231, the knockdown of c-Myc decreased LSD1 and TERT mRNA levels and LSD1 protein expression (Figs. 1C–F). These results suggest that c-Myc regulates the expression level of LSD1 in cancer cells. In AML, glioblastoma and prostate cancer, overexpression of c-Myc or LSD1 has already been reported.32,38–41) However, there is no report on whether both are overexpressed together. Therefore, we examined the relationship between LSD1 expression and c-Myc expression in various human tumors31–34) in The Cancer Genome Atlas (TCG A) database using cBioPortal.35,36) As shown in Fig. 1G, LSD1 mRNA expression correlated with c-Myc expression in human AML, glioblastoma, and prostate adenocarcinoma. These analyses support that LSD1 is potential novel c-Myc target gene. In addition, we also found that c-Myc and LSD1 are overexpressed together in stomach adenocarcinoma. These results suggested that c-Myc universally increases the expression of LSD1 in several human cancers.

(A, B) PC-3 cells were transfected with the indicated siRNAs for 72 h. (A) The expression of each gene was assessed by qPCR. The error bars indicated the standard deviation (S.D.). Significant differences are indicated as ** p < 0.01 and * p < 0.05. (B) Cell lysates were immunoblotted with the indicated antibodies. (C, D) HepG2 cells were transfected with the indicated siRNAs for 72 h. (C) The expression of each gene was assessed as in (A). (D) Cell lysates were immunoblotted with the indicated antibodies. (E, F) MDA-MB-231 cells were transfected with the indicated siRNAs for 72 h. (E) The expression of each gene was assessed as in (A). (F) Cell lysates were immunoblotted with the indicated antibodies. (G) Relationship between LSD1 expression and c-Myc in human tumor cells.31–34) Search results were analyzed by cBioPortal (http://www.cbioportal.org).35,36) Spearman’s correlation, and p values are noted on the right side of each graph.
To investigate the regulation of LSD1 expression by c-Myc, we prepared a human LSD1 promoter (−830 to +113)-luciferase reporter construct (Fig. 2A) and analyzed its responsiveness to c-Myc expression. c-Myc up-regulated the reporter activities of the LSD1 promoter (Fig. 2A) and artificial Myc-responsive promoter (6xMycRE-Luc) in COS-7 cells (Fig. 2B). c-Myc dimerizes with Max through a C-terminal bHLH-LZ domain to facilitate DNA binding.42) As shown in Fig. 2C, c-Myc ∆LZ failed to activate the promoter activity of LSD1. These results suggest that c-Myc activates LSD1 gene expression through its DNA binding.
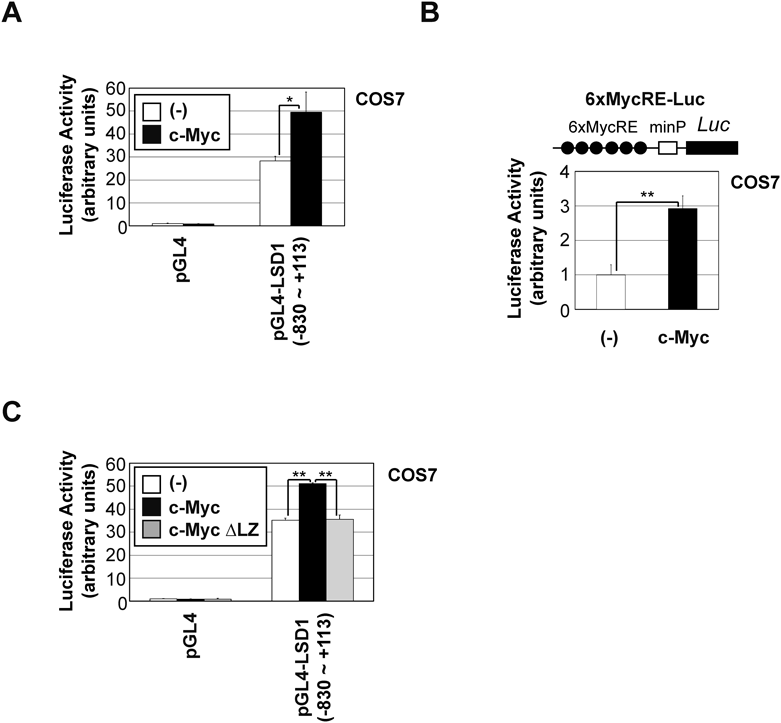
(A-C) COS7 cells were transfected with the indicated constructs. After 24 h, luciferase activity was measured. Experiments were performed in triplicate, and data are represented as mean-fold activation ± S.D. Significant differences are indicated as ** p < 0.01 and * p < 0.05.
To clarify the regulation of LSD1 induction via c-Myc expression, we identified the c-Myc responsive region in the LSD1 promoter using luciferase reporter plasmids of its promoter in various lengths, as shown in Fig. 3A. The reporter genes containing the region −110 to +28 were activated by c-Myc expression (Fig. 3B). This result indicates that c-Myc activates the LSD1 promoter via this region.
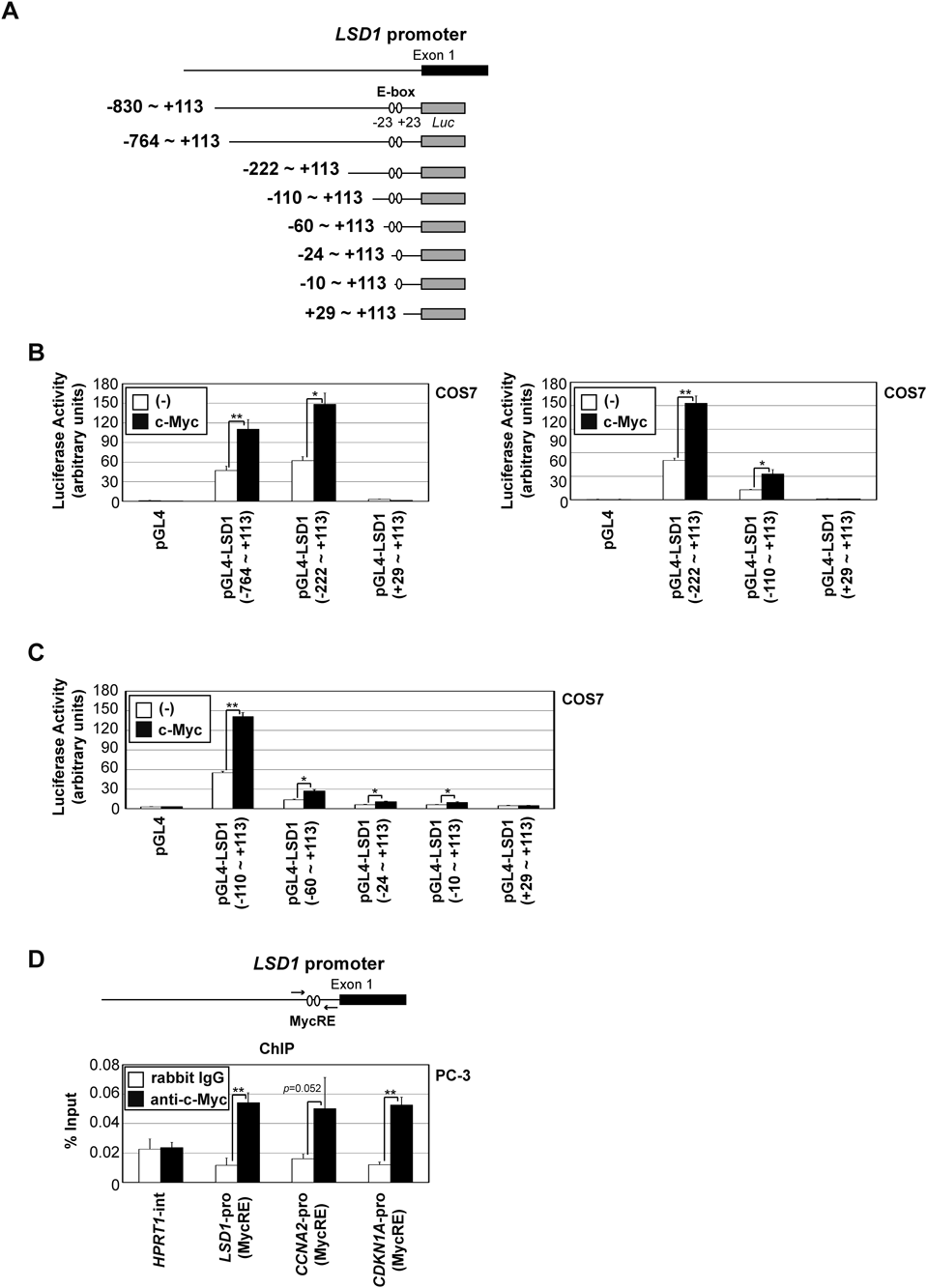
(A) A schematic representation of human LSD1 promoter constructs. (B, C) COS7 cells were transfected with the indicated constructs. After 24 h, luciferase activity was measured. Experiments were performed in triplicate, and data are represented as mean-fold activation ± S.D. (D) PC-3 cell lysates were subjected to a ChIP analysis with the indicated antibodies, and qPCR was conducted for the indicated promoters. Significant differences are indicated as ** p < 0.01 and * p < 0.05.
The c-Myc responsive region in the LSD1 promoter, −110 to +28, contained two non-canonical E-box sequences (−30 to −25: CTC GTG, +23 to +28: CGC GTG) (Fig. 3C). As shown in Fig. 3D, reporter plasmids carrying deletions of one or both non-canonical E-box sites showed minimal (−24 to +113, −10 to +113) and absent (+29 to +113) responses to c-Myc expression, respectively. Moreover, the ChIP analysis showed that c-Myc was detected in association with these E-box sites in the LSD1 promoter region, similar to the CCNA2 promoter and CDKN1A promoter29) (Fig. 3D).
In conclusion, we herein demonstrated that LSD1 is a downstream target of c-Myc. LSD1 may play a role in the oncogenic effects of c-Myc, such as the control of cancer stemness and malignant action of cancer (Fig. 4). A recent study reported that the combination of a BET inhibitor and LSD1 inhibitor exerted synergistic effects on the enhancement of apoptosis in a human AML xenograft model.43) The present study also supports the c-Myc-LSD1 axis being an attractive molecular target for cancer therapy.

Schematic representation of a mechanistic model for LSD1 regulation by c-Myc. In response to a mitogenic signal or Wnt/β-catenin signaling, c-Myc activity is induced. Activated c-Myc binds to the E-box sequences in the LSD1 promoter and activates the transcription of LSD1. The activation of c-Myc in cancer cells may be one of the mechanisms by which the aberrant up-regulation of LSD1 is induced.
The authors thank members of the Hayashi laboratory for their helpful discussions. We acknowledge the assistance of the Research Equipment Sharing Center at the Nagoya City University. This work was supported by a Grant-in-Aid for Scientific Research (C) (No. 15K07936, 15K07937, and 18K06660) from the Japan Society for the Promotion of Science (JSPS). YI was supported by the Aichi Cancer Research Foundation, the Foundation of Oriental Medicine Study, the Ichiro Kanehara Foundation, and the ONO Medical Research Foundation.
The authors declare no conflict of interest.