2019 Volume 42 Issue 5 Pages 850-855
2019 Volume 42 Issue 5 Pages 850-855
Acyl-CoA synthetase long-chain family members (ACSLs) are a family of enzymes that convert long-chain free fatty acids into their acyl-CoAs. ACSL4 is an ACSL isozyme with a strong preference for arachidonic acid (AA) and has been hypothesized to modulate the metabolic fates of AA. There are two ACSL4 splice variants: ACSL4V1, which is the more abundant transcript, and ACSL4V2, which is believed to be restricted to the brain. In the present study, we expressed recombinant human ACSL4V1 and V2 in Spodoptera frugiperda 9 (Sf9) cells using the baculovirus expression system and then partially purified both variants by cobalt affinity column chromatography. We then established a novel ACSL assay system with LC-MS/MS, which is highly sensitive and applicable to various kinds of fatty acids, and used it to investigate the substrate specificity of recombinant human ACSL4V1 and V2. The results showed that both ACSL4 variants preferred various kinds of highly unsaturated fatty acids (HUFAs), including docosahexaenoic acid (DHA), adrenic acid (docosatetraenoic acid) and eicosapentaenoic acid (EPA), as well as AA as a substrate. Moreover, our kinetic studies revealed that the two variants had similar relative affinities for AA, EPA and DHA but different reaction rates for each HUFA. These results confirmed the importance of both of ACSL4 variants in the maintenance of membrane phospholipids bearing HUFAs. Structural analysis of these variants might reveal the molecular mechanism by which they maintain membrane phospholipids bearing HUFAs.
Acyl-CoA synthetase long-chain family members (ACSLs) are a family of enzymes that convert long-chain free fatty acids into their acyl-CoAs.1,2) In humans and rodents, five ACSL isoforms have been identified: ACSL1, ACSL3, ACSL4, ACSL5, and ACSL6.2) Each ACSL isoform has a distinct substrate preference, subcellular localization and tissue distribution and has been suggested to be involved in the modulation of various pathophysiological events via the generation of long-chain acyl-CoA. Substrate specificity analysis using recombinant proteins expressed by Escherichia coli (E. coli) or COS7 cells revealed that among the ACSLs, ACSL4 (also known as ACS4 or FACL4) has a strong preference for arachidonic acid (AA).3,4) The substrate preference of this isoform suggests that ACSL4 is an important enzyme for controlling the levels of free AA. In fact, Golej et al. reported that overexpression of ACSL4 in human arterial smooth muscle cells markedly increased the synthesis of arachidonoyl-CoA, reduced cellular levels of unesterified AA, and blunted the secretion of prostaglandin (PG) E2.5) We also found that small interfering RNA-mediated knockdown of ACSL4 expression in rat fibroblastic 3Y1 cells significantly increased the release of AA and the production of its metabolites, including PGE2, PGD2 and PGF2α, compared with replicated control cells.6)
Although the substrate specificity of ACSL4 for fatty acids other than AA has not been fully elucidated, it has recently been shown that ACSL4 plays roles in adrenoyl and arachidonoyl esterification in the membrane, and thereby in ferroptosis, one of the types of programmed cell death.7,8) Ferroptosis is triggered by insufficiency of glutathione peroxidase 4 (GPx4), resulting in unchecked lipid peroxidation and eventual cell death. Suppression of long-chain polyunsaturated ω6 fatty acid incorporation into membrane phospholipids by genetic inhibition of ACSL4 makes it more difficult for membrane phospholipids to be peroxidized and then acts as a specific antiferroptotic rescue pathway.
ACSL4 is ubiquitously expressed but is especially abundant in the brain and the adrenal glands.3,9,10) Differential splicing generates two ACSL4 variants, ACSL4V1 and ACSL4V2.9,10) ACSL4V1 is the more abundant transcript and has been well-characterized, whereas ACSL4V2 appears to be restricted to the brain. ACSL4V2 contains an earlier in-frame start codon, which translates into an additional 41 amino acids. Since a point mutation of the ACSL4 gene affecting both transcripts is one of the genetic causes of non-specific X-chromosome-linked mental retardation,10,11) ACSL4V2 is suggested to play an important role in neural development. However, the enzymatic characteristics of ACSL4V2 remains unclear.
In the present study, to investigate the enzymatic characteristics of ACSL4V1 and V2 including their substrate specificity for various kinds of fatty acids, we cloned the cDNAs for human ACSL4V1 and V2 and then expressed recombinant enzymes containing a C-terminal His-tag in Spodoptera frugiperda 9 (Sf9) cells using the baculovirus expression system. In addition, we established an ACSL assay system using LC-MS/MS, which is highly sensitive and applicable to various kinds of fatty acids, and examined the substrate specificity of the recombinant human ACSL4V1 and V2 proteins.
RNA obtained from human Caco-2 cells was subjected to reverse transcription using SuperScript III reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, U.S.A.). The cDNA encoding human ACSL4 with a C-terminal His(6) tag was prepared as follows: the sense primer for V1 (5′-CAC C ATG GCA AAG AGA ATA AAA GCT AAG C-3′), sense primer for V2 (5′-CAC C ATG AAA CTT AAG CTA AAT GTG CTC AC-3′) and antisense primer (5′-TTA ATG GTG ATG GTG ATG ATG TTT GCC CCC ATA CAT TCG T-3′) were paired to amplify cDNA products from the human Caco-2-derived cDNA by PCR (35 cycles at 94°C for 10 s and 68°C for 2 min) using high-fidelity KOD plus DNA polymerase ver. 2 (Toyobo, Osaka, Japan). The PCR products were further subjected to PCR with A-attachment mix (Toyobo) for 10 min at 60°C to add an adenosine base to the 3′-ends on both the strands, followed by subcloning into a pTAKN-2 vector (BioDynamics Laboratory Inc., Tokyo, Japan). The plasmids were digested with BamHI/XhoI, and the resulting inserts were ligated into a pFASTBac1 vector (Invitrogen, Carlsbad, CA, U.S.A.) at the BamHI/XhoI sites. The plasmids were used for bacmid preparation using the Bac-to-Bac Baculovirus Expression System (Thermo Fisher Scientific) according to the manufacturer’s instruction, and the purified bacmids were then transfected into Sf9 cells with Cellfectin reagent (Invitrogen) to produce a baculovirus bearing the C-terminally His(6)-tagged ACSL4. Amplification of the virus was performed by adding an aliquot of the initial virus pool to fresh, sub-confluent Sf9 cells in 150-mm dishes. After incubation for 5 d at 27°C, all the infected cells were pelleted by centrifugation, washed with phosphate buffered saline (PBS), resuspended in 20 mM phosphate buffer pH 7.0 containing 500 mM NaCl (buffer A), sonicated (4 × 2 min bursts) using a BRANSON sonicator at 30% output, and centrifuged at 10000 × g for 5 min. Then, recombinant ACSL4 variant proteins were partially purified using a TALON column (GE Helthcare, Uppsala, Sweden) as previously described.12,13) The 10000 × g supernatant was applied onto a 4 mL TALON column equilibrated with buffer A and then washed with buffer A and 10 mM imidazole-containing buffer A. The partially purified recombinant ACSL4 variants were eluted with 100 mM imidazole-containing buffer A, and used as an enzyme source for the ACSL enzyme assay.
Measurement of ACSL ActivityEnzyme preparations (10 µg) were incubated with 0.1–10 nmol of the indicated fatty acids in 100 µL assay buffer (50 mM phosphate buffer pH 7.0, 1% Triton-X 100, 5 mM dithiothreitol, 15 mM MgCl2, 1 mM CoA, 10 mM ATP) for 20 min at 37°C. The reaction was terminated by addition of 1 mL of methanol, and spiked with an internal standard (5 pmol heptadecanoyl-CoA, 17 : 0-CoA). Following centrifugation at 20000 × g for 5 min, the supernatants were dried up by a centrifugal evaporator, resuspended in 100 µL of methanol, and centrifuged at 20000 × g for 5 min; the resulting supernatants were analyzed by LC-MS/MS to quantify acyl-CoA species.
Electrospray Ionization Mass Spectrometry AnalysisAll mass spectrometric analyses were performed using a Prominence HPLC system (Shimadzu, Kyoto, Japan) equipped with a linear ion trap quadrupole mass spectrometer (QTRAP5500; Sciex, Framingham, MA, U.S.A.). Quantification of long-chain fatty acyl-CoA species was performed using LC-MS/MS via multiple-reaction monitoring (MRM) in positive-ion mode by a modified protocol of acyl-CoA analysis as previously reported.14) Briefly, 5 µL of sample was injected into a Gemini C18 column (2.0 × 150 mm, 5.0 µm, 110 Å; Phenomenex, Torrance, CA, U.S.A.). The LC flow was maintained at 0.2 mL/min, and 1) started with 80% mobile phase A (1 mM ammonium bicarbonate pH 8.3) and 20% mobile phase B (acetonitrile) for the initial 3 min; then 2) the mobile phase B was increased to 100% within 8 min and maintained at 100% for 15.5 min; and finally 3) the gradient was finally returned to the initial condition with in 0.5 min and the column was re-equilibrated at the initial conditions for 5 min for the next injection. The neutral loss of m/z 507 was used to detect each long-chain fatty acyl-CoA species. Five picomoles of 17 : 0 CoA was added to samples to quantify various acyl-CoA species by comparing the relative peak areas in the ion chromatogram in the MRM mode.
We first cloned the cDNA for human ACSL4 variants, ACSL4V1 and ACSL4V2, from human epithelial colorectal adenocarcinoma Caco-2 cells. It has been reported that the expression of ACSL4 is up-regulated in colon adenocarcinoma.15) Although it has been reported that ACSL4V2 may be restricted to the brain in normal tissues,9,10) Caco-2 cells express both ACSL4 variants, ACSL4V1 and ACSL4V2. Plasmids containing the cDNAs for ACSL4V1 and ACSL4V2 were used for bacmid preparation; then recombinant ACSL4 variants with a C-terminal His-tag were expressed in Sf9 cells. It has been reported that both ACSL4 variants are membrane-associated enzymes. Küch et al. showed that ACSL4V1 localized to the inner side of the plasma membrane as well as in the cytosol of HepG2 cells, and that ACSL4V2 which contains an additional N-terminal hydrophobic region was localized at the endoplasmic reticulum and on lipid droplets.16) Moreover, several prediction programs of transmembrane helices in proteins such as TMHMM17) revealed that the membrane spanning region is not predicted in ACSL4V1, but the relatively high score is predicted in the N-terminal extension of ACSLV2. However, our immunoblot analysis revealed that both ACSL4 variants were detected in 10000 × g supernatants as well as 10000 × g pellets of ACSL4-expressing Sf9 cell lysates (Fig. 1). These results suggested that both ACSL4 variants may be peripheral membrane proteins, although the mechanism how the signal recognition particle avoids the hydrophobic region of ACSL4V2 remain to be elucidated. Further studies will be needed to clarify their membrane topologies.
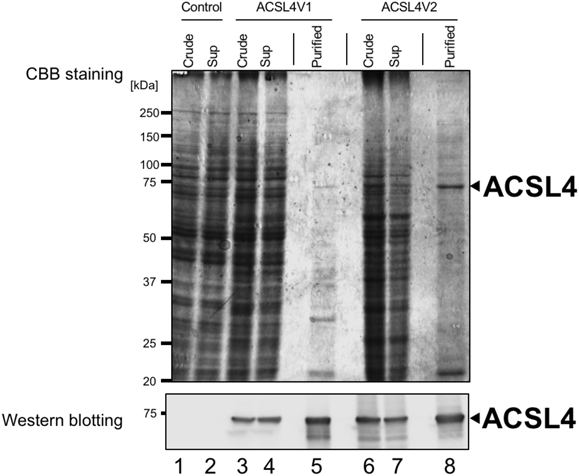
Sf9 cells grown in 100-mm dishes were infected with baculovirus bearing the C-terminally His(6)-tagged ACSL4V1 or ACSL4V2 for 3 d. The expression of ACSL4 proteins were assessed by CBB staining (upper panel) and Western blotting (lower panel). Lanes 1 and 2 show 15 µg of uninfected Sf9 cell lysate and 10000 × g supernatant. Lanes 3 and 4 show 15 µg of ACSL4V1 baculovirus-infected Sf9 cell lysate and 10000 × g supernatant. Lanes 6 and 7 show 15 µg of ACSL4V2 baculovirus-infected Sf9 cell lysate and 10000 × g supernatant. Lane 5 band 8 shows imidazole-eluted fractions containing partially purified ACSL4V1 and V2, respectively. Anti-ACSL4 antibody (Rb mAb to FACL4 (ab155282); Abcam, Cambridge, U.K.). was used for Western blotting.
In the present study, we used 10000 × g supernatants of ACSL4-expressing Sf9 cell lysates as enzyme sources of ACSL4V1 and V2, and partially purified these ACSL4 variants by cobalt affinity column chromatography. As shown in Fig. 1, recombinant ACSL4 variants bound to a TALON column were eluted with 100 mM imidazole-containing buffer. Both of partially purified ACSL4 variants showed molecular weights of about 75 kDa, and as expected, the molecular weight of ACSL4V2 was slightly greater than that of ACSL4V1. We further investigated ACSL activities of the recombinant ACSL4 variants using these partially purified enzymes.
Establishment of a Novel ACSL Assay System Using LC-MS/MSUntil now, ACSL activity has mainly been assayed using radioactive substrates5,11,18) or a spectrophotometric assay method.3,19–21) When radioactive fatty acids are used as substrates of ACSL, ACSL activity is measurable with high sensitivity. However, there is a limit to the variety of commercially available radioactive fatty acids. In the spectrophotometric assay method, ACSL activity is assayed by coupling the reaction of ACSL with those of adenylate kinase, pyruvate kinase and lactate dehydrogenase and quantifying the oxidation of reduced nicotinamide adenine dinucleotide (NADH) at 334 nm. This method is applicable to various kinds of fatty acids, but it is an indirect assay and has a slightly lower sensitivity, with a detection limit at the nanomolar level. Thus, we here established a novel ACSL assay system using LC-MS/MS, which is highly sensitive and applicable to various kinds of fatty acids.
Li et al. developed a programmed multiple reaction monitoring (MRM) method in LC/MS-MS to quantitate acyl-CoAs in various rat organs.14) In positive electrospray ionization-MS/MS, fragments of acyl-CoAs from the neutral loss of 507 were the most abundant fragment; thus, this fragment was used for acyl-CoA MRM. We incubated the partially purified ACSL4V1 and V2 proteins with various kinds of fatty acids, and then quantitatively scanned acyl-CoAs in enzymatic products according to the characteristic fragmentation pattern of acyl-CoAs in MS/MS. It has been shown that E. coli-expressed recombinant ACSL4V1 has a strong preference for AA and eicosapentaenoic acid (EPA).3) As shown in Fig. 2, our assay system using LC-MS/MS confirmed that recombinant human ACSL4V1 protein converted AA (20 : 4) and EPA (20 : 5) to arachidonoyl-CoA (20 : 4-CoA) and eicosapentaenoyl-CoA (20 : 5-CoA), respectively. Conversion of docosahexaenoic acid (22 : 6) and adrenic acid (22 : 4) to their acyl-CoAs by ACSLV1 was also observed in our system. Likewise, recombinant human ACSL4V2 also acted on these highly unsaturated fatty acids (HUFAs), converting them to their respective acyl-CoAs. There seemed to be no differences in substrate specificity between ACSL4V1 and V2. Overall, the results confirmed the importance of ACSL4 in the maintenance of membrane phospholipid bearing HUFAs. The two ACSL4 variants might perform similar functions in cellular organelles of tissues that express them differentially.

Various kinds of fatty acids (solid line) or vehicles (dashed line) were incubated with the partially purified recombinant ACSL4V1 (A) or ACSL4V2 (B), and then acyl-CoAs in enzymatic products were quantitatively scanned according to the characteristic fragmentation pattern of acyl-CoAs in MS/MS. MRM chromatograms are shown.
Since 20 : 4-CoA, 20 : 5-CoA and 22 : 6-CoA are commercially available, their use as standards enabled us to perform quantitative analysis of the kinetics of ACSL4V1 and V2 on AA, EPA and docosahexaenoic acid (DHA). The substrate concentrations of AA, EPA and DHA were varied (1–100 µM), and then substrate–velocity curves and linear Lineweaver-Burk plots on these three fatty acids were observed for ACSL4V1 and V2. As shown in Fig. 3 and Table 1, ACSL4V1 and ACSL4V2 showed similar Km values for AA, and both variants had similar affinities for AA, EPA and DHA. On the other hand, both variants’ reaction rate for the various HUFAs were different, with the Vmax for AA being the fastest. The Vmax values of both variants were in the order of AA > EPA > DHA, although previous reports described that the Vmax value for EPA of E. coli-expressed recombinant ACSL4V1 was rarely different from that for AA.3) Klett et al. further identified differences in substrate preference for epoxyeicosatrienoic acids (EETs) and hydroxy EETs (HETEs) between bacterial and mammalian expressed ACSL isoforms.4) Baculovirus-expressed recombinant enzymes are more similar in structure to mammalian recombinant enzymes than to bacterial ones. Our data indicated that ACSL4 plays an especially important role in arachidonate metabolism. Structural analysis of these variants might reveal the molecular mechanism by which they maintain membrane phospholipid bearing HUFAs.

ACSL assays using the partially purified recombinant ACSL4V1 (A) or ACSL4V2 (B) (10 µg) were performed under different concentrations (1–100 µM) of AA (●), EPA (△) and DHA (■). Data points represent the means of three determinations obtained for each experiment.
| Variants | Substrates | Km (µM) | Vmax (nmol/min/mg) | Vmax/Km (mL/min/mg) |
|---|---|---|---|---|
| ACSL4V1 | AA | 9.80 ± 2.08 | 6.88 ± 2.47 | 0.702 |
| EPA | 5.31 ± 2.56 | 0.73 ± 0.18 | 0.137 | |
| DHA | 5.45 ± 0.60 | 0.29 ± 0.09 | 0.053 | |
| ACSL4V2 | AA | 7.02 ± 3.78 | 4.87 ± 1.65 | 0.694 |
| EPA | 13.30 ± 1.19 | 1.21 ± 0.59 | 0.091 | |
| DHA | 14.90 ± 2.71 | 0.47 ± 0.15 | 0.032 |
Km and Vmax value were estimated by linear Lineweaver–Burk plot as shown in Fig. 3.
This work was supported in part by a Grants-in-Aid for Scientific Research (B) (No. 16H05108) from the Japan Society for the Promotion of Science, and by the Private University Research Branding Project from the Ministry of Education, Sports, Science, Culture and Technology of Japan.
The authors declare no conflict of interest.