2019 Volume 42 Issue 6 Pages 867-872
2019 Volume 42 Issue 6 Pages 867-872
Dynamic conformational transitions and molecular assemblies are essential properties of proteins, and relevant to their biological and pathological functions. Neurodegenerative diseases are known to be caused by abnormal, toxic assemblies of related proteins, e.g., amyloid β (Aβ) in Alzheimer’s disease. Growing evidence indicates that the aggregation of various amyloidogenic proteins, including Aβ, can be highly enhanced at glycolipid membranes, suggesting that dynamic glycolipid-dependent conformational changes of proteins constitute crucial steps for their subsequent pathogenic amyloid fibril formation. It has also been proposed that several proteins, including molecular chaperones, can capture amyloidogenic proteins and thereby suppress their fibrillization. NMR spectroscopy provides a powerful tool for characterizing the conformational dynamics and intermolecular interactions of proteins, as well as for exploring transiently formed weak interactions among proteins in solution with various biomolecules, such as glycolipids. Our research group therefore attempted to elucidate the structural basis of protein–glycolipid and protein–protein interactions that either promote or suppress molecular assemblies of amyloidogenic proteins, using both solution and solid-state NMR methods in conjunction with other biophysical techniques. Our findings provide structural views of molecular processes involving amyloidogenic proteins of clinical and pathological interest and offer clues for the development of drugs to prevent and treat neurodegenerative diseases.
Proteins are the most versatile biomacromolecules in living systems and have many crucial functions in various biological processes. The three-dimensional structure of a protein plays an important role in the performance of its biological function. In addition to X-ray crystallography, solution and solid-state NMR spectroscopic approaches are powerful ways to obtain structural information about proteins at an atomic level of resolution. The principal behind NMR-based structure determination is to obtain a set of empirical structural parameters, e.g., chemical shift, scalar J-coupling, dipolar coupling, and nuclear Overhauser effect (NOE).1) Dynamic conformational change is another essential property of proteins, which is required to regulate their biologically functional processes as typified by molecular recognition. The motional freedom of a protein provides conformational plasticity that is indispensable during interaction and reaction processes, and also for the conformational adaptability of proteins to their binding partners. For example, conformational ensembles of unbound proteins are converged to their selected and stabilized states on binding to their cognate ligands.2–4)
NMR spectroscopy is useful for characterizing the conformational dynamics and intermolecular interactions of proteins over a wide range of time scales.5–8) For instance, a longitudinal rate (R1), a transverse relaxation rate (R2), and a heteronuclear NOE can provide information on conformational fluctuations at the subnanosecond time scale. Protein backbone dynamics have been delineated with order parameters and correlation times of individual amino acid residues by a model-free formalism based on measurements of 15N R1, R2, and {1H}-15N NOE.
NMR relaxation dispersion experiments have characterized conformational interconversion of biomolecules occurring on the microsecond to millisecond time scale, quantifying chemical shifts, conformer populations, and exchange rates. Residual dipolar coupling (RDC) is sensitive to protein motion that is either faster or slower than its rotational correlation time, giving order parameters describing protein motions occurring over time scales ranging from picoseconds to milliseconds. H/D-exchange experiments can probe slow dynamics of proteins occurring over time scales of seconds or longer, to characterize local conformational stability and global unfolding of protein structures.
NMR spectroscopy is also uniquely suitable for observing transiently formed weak interactions in solution between proteins and other molecules, such as other proteins, lipids, and other chemical compounds.9) In particular, NMR has been used to delineate the coupled binding and folding processes of intrinsically disordered proteins that lack stable secondary or tertiary structures, and often become structured on interacting with their binding partners during molecular recognition processes.10,11) In structure-based drug design programs, NMR screening has been used to detect weak binding of compounds to target proteins.12) Thus NMR has contributed not only to an understanding of the mechanisms underlying protein function but also to drug discovery research.
It has been reported that misfolded and aggregated proteins can play a pathological role in cell dysfunction and tissue damage and may lead to the so-called “protein conformational diseases,” including amyloidosis, type II diabetes, and prion diseases.13) A characteristic feature of these diseases is they can give rise to the dynamic conformational conversion of native proteins into the form of insoluble amyloid fibrils, in which multiple layers of β-sheets are regularly stacked.14,15) Certain neurodegenerative diseases are also known to be caused by abnormal, toxic assemblies of related proteins, e.g., amyloid β (Aβ) in Alzheimer’s disease (AD) and α-synuclein (αSN) in Parkinson’s disease.
Growing evidence indicates that the aggregation of various amyloidogenic proteins such as Aβ and αSN can be highly enhanced at glycolipid membranes, suggesting that dynamical glycolipid-dependent conformational changes in proteins are a crucial step for the subsequent amyloid fibril formation that may lead to neurodegenerative disease.16–20) However, no detailed structural study has reported on how proteins bind to glycolipid membranes. Conformational analysis of protein–glycolipid complexes is one of the most challenging subjects for NMR spectroscopy, because such complexes are relatively huge and unstable, with high fluidity and flexibility. Additionally, the interplay among proteins, oligosaccharides, and lipids is characterized as nonstoichiometric and heterogeneous in nature. Another factor that has prevented detailed structural analysis of such systems is that proteins interacting with glycolipids swiftly become fibrotic, making it difficult to observe any unstable intermediates of protein–glycolipid complexes. Recently, it has been proposed that molecular chaperones actively contribute to the suppression of toxic aggregate formation of various proteins associated with neurodegenerative disorders.13,21–23) However, the mechanisms underlying chaperone-mediated inhibition of these pathological molecular processes remain unclear.
Given the factors outlined, we attempted to overcome these difficulties by using both solution and solid-state NMR methods combined with other biophysical techniques so as to provide a structural basis for protein–glycolipid and protein–protein interactions that are associated with molecular mechanisms by which amyloid formation may be promoted or suppressed.
Aβ is usually a 40- or 42-amino-acid peptide cleaved from its precursor membrane protein by the sequential actions of β- and γ-secretases. AD is associated with progressive accumulation of Aβ deposits in the brain resulting from formation of extracellular senile plaques.24,25) In aqueous solution, Aβ exists mostly as a monomeric unstructured polypeptide, although molecular dynamics (MD) simulations identified local non-random conformations in Aβ molecules.26,27) A paramagnetism-assisted NMR technique has been developed for characterizing the dynamic conformational ensemble and the transient intra-molecular interactions of Aβ in solution.28) This technique exploits paramagnetic probes such as spin labels attached to specific positions in Aβ molecules, which can induce paramagnetic relaxation enhancement (PRE) effects as NOE-independent sources of long-distance information. The conversion of soluble, nontoxic, monomeric Aβ to its toxic, aggregated form, like oligomers or fibrils, has been suggested to be an extremely important step in AD progression. PRE-measurements have also been applied to elucidate the intermolecular interactions of Aβ oligomers. Recently, NMR-derived high-resolution structural models have visualized Aβ(1–42) fibrils adopted an S-shaped conformation forming a salt bridge between Lys28 and the C-terminal carboxylate, while Aβ(1–40) fibrils assume U-shaped conformation.15,29–34)
Yanagisawa et al. have previously reported that a unique Aβ species tightly associated with GM1 ganglioside was identified in cerebral cortices from AD patients.35) GM1 is a glycosphingolipid abundant in neuronal membranes, possessing a pentasaccharide including a sialic acid, which exhibits a bouquet-like structure that is capable of mobility.36–39) Based on a series of in vivo and in vitro studies demonstrating that GM1-bound Aβ accelerates Aβ assembly,17,20,40) it has been proposed that this Aβ species acts as a seed for cerebral Aβ fibril formation. Furthermore, a GM1-induced toxic Aβ(1–40) assembly has been reported enhanced in Arctic-type Aβ, one of hereditary Aβ variants.41) It is also notable that Aβ(1–40) interacting with GM1 micelles forms fibrils with higher toxicity.42) These findings prompted us to perform structural characterization of GM1-bound Aβ for better understanding of the molecular mechanism underlying the onset of AD.43) In this context, we carried out a series of NMR studies for characterizing the interactions of Aβ with GM1 clusters by employing appropriate membrane models and have successfully provided a structural basis for this pathogenic interaction, as shown in Fig. 1.
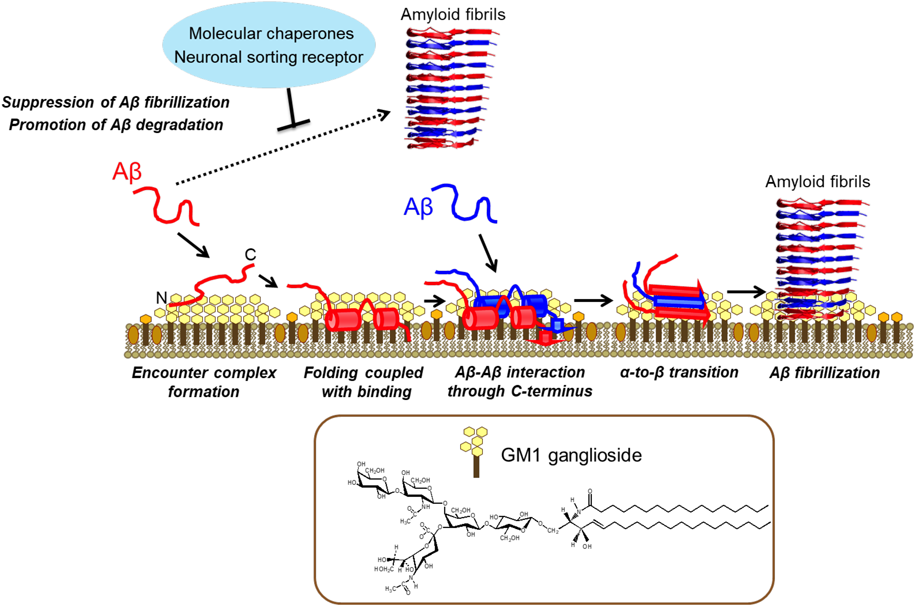
The GM1 cluster captures Aβ at the hydrophobic/hydrophilic interface, which facilitates α-helix formation, thereby restricting the spatial rearrangements of Aβ molecules. Consequently, a specific intermolecular interaction between Aβ molecules is enhanced on the GM1 cluster leading to their α-to-β conformational transition resulting in amyloid fibril formation. Several proteins, including molecular chaperones, capture Aβ and thereby suppress its fibrillization. (Color figure can be accessed in the online version.)
The chemical shift perturbation and PRE data highlighted the role of the sugar–lipid interface of the ganglioside clusters in accommodating Aβ.36) The saturation transfer data based on transverse relaxation-optimized spectroscopy and chemical shift titration indicate that the interaction between GM1 micelles and Aβ molecules involves a process of accommodation for the two α-helices, His14-Val24 and Ile31-Val36, plus the C-terminal Val39-Val40 dipeptide segment, in the hydrophobic interior.36,44,45) It has been hypothesized that this restricted topology facilitates oligomerization of Aβ molecules at the sugar–lipid interface in membranous environments. Indeed, in the MD simulation validated by PRE-assisted NMR experiments, Aβ(1–40) exhibited a hairpin structure at the interface more readily than in bulk water, indicating that the formation of intermolecular β-sheet structures can be accelerated more easily at the interface compared with formation in bulk water.46) Furthermore, our solution NMR data suggest that Aβ adopts a β-structure at higher densities on the clusters of GM1, whereas an α-to-β conformational transition on 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) lipid bilayers was revealed by our solid-state NMR measurements.47,48)
These findings demonstrate that the interaction between Aβ(1–40) and GM1 clusters involves multiple steps, including accommodation, conformational transition, and subsequent oligomerization processes. Namely, the GM1 cluster provides a unique platform at its hydrophobic/hydrophilic interface for binding coupled with the α-helix formation of Aβ molecules. This restricts the spatial rearrangements of Aβ molecules and promotes a specific intermolecular interaction and α-to-β conformational transition leading to the formation of amyloid fibrils (Fig. 1).
Hereditary mutations have potential impact on these on-membrane molecular events49) as exemplified by the Flemish-type mutation (A21G), which disrupts the first α-helix identified in wild-type Aβ(1–40) bound to lyso-GM1 micelles, rendering the unstructured N-terminal segment tethered through the residual C-terminal helix.50) This mutation prevents the initial nucleation process of Aβ(1–40) fibril formation, suggesting that it perturbs the membrane binding properties of Aβ and thereby affects its nucleation behavior during fibrillization.
It has been reported that molecular chaperones actively contribute to the suppression of toxic aggregate formation of various amyloidogenic proteins associated with neurodegenerative disorders.13,21–23) It has also been reported that various molecular chaperones, such as heat shock proteins, prefoldins, and small heat shock proteins, are recognized to inhibit protein aggregation and mediate protein degradation via the ubiquitin-proteasome system or autophagy.51)
In particular, protein disulfide isomerase (PDI), an essential folding catalyst and important player in protein quality control in the endoplasmic reticulum, is upregulated in the brain of patients with Parkinson’s disease.52) This protein is found in Lewy bodies, which also contain α-synuclein (α-SN), an intrinsically unstructured protein consisting of 140 amino acid residues. Increased expression of PDI was also observed in αSN transgenic mice.53) Intriguingly, the specific hydrophobic segment of αSN, which was identified by X-ray crystallography in conjunction with NMR-based validation, is capable of interacting with the eukaryotic chaperone PDI,54) the bacterial chaperone GroEL,55) and the archaeal chaperone PbaB.56) These observations suggest that αSN displays a chaperone-philic binding motif that can be widely recognized as a mimic of misfolded protein hallmarks. NMR data also indicate that αSN, as well as Aβ, when non-covalently tethered to GroEL remains largely unfolded and highly mobile, enabling identification of the interaction hot spots displayed on these disordered proteins.55,57,58)
The interaction between hydrophobic segments of amyloidogenic proteins and molecular chaperones as revealed by NMR could be a key feature in preventing protein aggregation. For instance, GroEL could suppress Aβ(1–40) amyloid formation by transiently interacting with the two hydrophobic segments Leu17-Ala21 and Ala30-Val36 of Aβ(1–40), which involve key residues in fibril formation.48,58) Such dynamic but loose complexes have been observed for a variety of interaction systems that contribute to the specific suppression of toxic amyloid formation by neurodegenerative disordered proteins, such as a neuronal sorting receptor SorLA, which captures Aβ inside a tunnel to extend the β-sheet of one of its propeller blades.59) In conjunction with X-ray crystallography, NMR spectroscopy demonstrated that Aβ can remain attached to SorLA while undergoing transitions among different bound states involving multiple capture sequences, suggesting that SorLA binds Aβ monomers through weak interactions and escorts them to lysosomes for degradation.59)
These results provide deeper insights into the underlying mechanisms of the molecular recognition of amyloidogenic proteins of pathological interest.
Our findings demonstrate that ganglioside clusters offer dynamic environments for the conformational transition, spatial rearrangement, and transient intermolecular interaction of amyloidogenic proteins. The observed conformational multiplicities, along with lower affinities in the protein–carbohydrate interaction systems, are unique but difficult properties to study using both experimental and theoretical approaches. To describe the other transiently interacting steps such as the initial encounter complex formation, it is necessary to use appropriately designed ganglioside clusters. This is because it is conceivable that the size and curvature of ganglioside clusters are determining factors of their interactions with proteins, leading to consequent oligomerization and fibrillization events. Self-assembled spherical supramolecular complexes displaying an oligosaccharide cluster,60) as well as ganglioside-embedding small bicelles,61) have been recently proposed as intriguing examples of nanoscale standardized stages for detailed NMR characterization of carbohydrate–protein interactions taking place on glycolipid clusters, giving insights into the ganglioside-binding specificity of Aβ during the initial encounter complex formation.
Another important issue is the direct observation of transient Aβ–Aβ interactions in the elongation phase of fibril formation. NMR techniques such as relaxation dispersion and saturation transfer experiments, which explore invisible NMR states, are useful approaches for indirect observation of the transient conformation of monomeric Aβ bound to large fibrils and oligomers.62,63) In addition, it is important to provide clues for the design and development of novel drugs to antagonize Aβ fibrilization based on the detailed structural characterization of the putative conformational epitope at the growing end of its fibril. An MD simulation based on the NMR-derived structural model of an Aβ fibril has indicated that the Aβ at the growing end of an amyloid fibril adopts a β-hairpin conformation with less fluctuation, as compared with the flexible opposing terminus.64) Moreover, because various environmental factors can dramatically influence fibril structures and aggregation kinetics, it is essential to perform structural analyses with conformational trapping by applying high pressure, microgravity, point mutations, pH optimization, covalent constraints, and so on. To address these issues, it is inevitable to take a multilateral approach with appropriately designed model systems using NMR spectroscopy in conjunction with other advanced biophysical methods, such as solution scattering, electron microscopy, and high-speed atomic force microscopy.65–67) Integration of these biophysics-based experimental and theoretical approaches will provide important clues for quantitative understanding of the molecular processes involved in the onset and development of amyloid diseases. Moreover, it will give fundamental insights into the universal molecular mechanisms that govern amyloid formation across diverse systems, leading to new concepts for designing drugs that target neurodegenerative diseases. Accordingly, research targeting “amyloids” inspires us from the point of view of pharmaceutical applications as well as basic molecular science.
I would like to express my sincere gratitude to Prof. Koichi Kato (ExCELLS and IMS) for his mentorship and to Dr. Katsuhiko Yanagisawa (National Center for Geriatrics and Gerontology) and Prof. Christopher M. Dobson (University of Cambridge) for their great guidance and suggestions. I would like to acknowledge Prof. Makoto Fujita (University of Tokyo), Dr. Yoshiki Yamaguchi (RIKEN), Dr. Katsuyuki Nishimura (IMS), and Dr. Sota Sato (University of Tokyo) for their precious collaborations. I also sincerely thank all the collaborators and all the laboratory members for sharing fruitful research.
The author declares no conflict of interest.
This review of the author’s work was written by the author upon receiving the 2018 Pharmaceutical Society of Japan Award for Young Scientists.