2020 Volume 43 Issue 2 Pages 306-318
2020 Volume 43 Issue 2 Pages 306-318
This study focused on the differential metabolomic effects between water extracts of Polygoni Multiflori Radix and Polygoni Multiflori Radix Preparata in rats. The extracts were subsequently administered for 28 d. Serum biochemical indicators were tested, hematoxylin–eosin staining and immunohistochemistry staining were used to detect histopathological changes in the livers. Ultra-performance LC/quadrupole time-of-flight mass spectrometry was used to detect the changes in endogenous metabolites. Finally, we performed detailed analysis of the changes in metabolic pathways. Hematoxylin–eosin staining and immunohistochemistry staining results indicated that the water extracts of Polygoni Multiflori Radix and Polygoni Multiflori Radix Preparata had mild liver injury effect. Fifty-two differential endogenous biomarkers were confirmed as potential biomarkers between Polygoni Multiflori Radix and Polygoni Multiflori Radix Preparata groups. In the positive ion mode, the biomarkers included 31 Phosphatidyl cholines (PCs), six lysoPCs, and ceramide. In the negative ion mode, 12 biomarkers were confirmed, including glycodeoxycholic acid, chenodeoxycholic acid, and deoxycholic acid, etc. In Hydrophilic Interaction Liquid Chromatography (HILIC) mode, nine biomarkers were confirmed, including niacinamide, L-palmitoylcarnitine, and butyrylcarnitine, etc. Using MetaboAnalyst 4.0, six related metabolic pathways, including taurine and hypotaurine metabolism, sphingolipid metabolism, glycerophospholipid metabolism, nicotinate and nicotinamide metabolism, arginine and proline metabolism, and tryptophan metabolism and primary bile synthesis, were confirmed as the most differential pathways between the Polygoni Multiflori Radix and Polygoni Multiflori Radix Preparata groups.
Polygoni Multiflori Radix (PMR) and Polygoni Multiflori Radix Preparata (PMRP) have been used as traditional Chinese Medicine for centuries throughout the world.1) They are two forms of clinical preparation of Polygonum multiflorum Thunb.2) Due to the processing method, they display marked differences in clinical use. PMR is used as an antioxidant and for purgation, whereas PMRP is used as a tonic and an antiaging drug.3) The pharmacological effects of PMR are detoxification, elimination, malarial treatment, and laxative, whereas PMRP has been shown to have positive effects on the liver and kidneys, as well as display benefits for the blood, anticancer, hepatoprotective and immunomodulation effects, turbidity and lipid lowering anti-Alzheimer’s disease activity.4–11)
Increasing reports of drug induced liver damage (DILI) caused by traditional Chinese medicine (TCM) have been reported in recent years, with many of these reports being attributed to systemic liver toxicity.12–14) As a commonly used tonic with a wide range of applications, these frequently reported adverse reactions caused by PMR or PMRP have become a major issue.15–18) PMR and its prescriptions can lead to pronounced liver damage. Official documents attributed to the medicine include relevant warning information and the correction scope of prescription. Animal studies have indicated that PMR has a significant interference effect on the bile acids composition of rats.19) PMR-induced liver damage is mainly due to its effect on CYP7A1, a key enzyme in bile acid metabolism.20) Rats treated with PMR for seven days did not experience liver damage, but did show accelerated bile acid enterohepatic circulation. This, in turn, changed the composition of intestinal bile acids, leading to the activation of an fxr-fgf15 signal in intestines, which can inhibit the expression of CYP7A1 in the liver.21)
DILI has occurred in only a small number of individuals when taking other medicines. These patients all presented with the typical characteristics of idiosyncratic drug-induced liver injury.22,23) The level of cis-stilbene glycoside was significantly increased in most PMR samples, which induced liver injury in patients. It was also observed that that cis-stilbene glycoside positively correlated with PMR-induced liver injury.15,24) The study demonstrated that PMR and PMRP do not induce liver damage as judged from cell proliferation, the integrity of cell membranes, and overall enzyme secretion in vitro. Moreover, three major chemical constituents of PMR, stilbene glycoside, physcion, and emodin showed no clear cytotoxicity against the human liver cell line L02.15) Chemical constituents–cytotoxicity relationship investigations revealed that stilbene glycoside and physcion had an attenuating effect on emodin. The processing of PMR may reduce its effect on cell proliferation and enzyme secretion in liver cells.15) Based on this, it is necessary to study liver damage caused by PMR and PMRP.
Metabolomics is a new approach for comprehensively profiling small molecules that can be used to detect and semi-quantitatively measure the levels of hundreds of unique metabolites from a broad range of metabolic pathways.25) In agreement with the holistic concept of TCM, metabolomics has shown great potential in efcacy and toxicity evaluation of TCM. A major aim of metabolomics is to obtain answers to provide insight into biological questions. Metabolomics is suitable for revealing the biochemical changes of disease pathogenesis.26,27) In this study, we systematically studied the differential metabolism effects of water extracts between PMR and PMRP on rats. We intended to discover endogenous differential metabolites and metabolic pathways, while analyzing the functions of these biomarkers and metabolic pathways.
Methanol, acetonitrile, formic acid, and ammonium acetate were purchased from Sigma-Aldrich (Shanghai, China). Water was available from a Milli-Q water purification system (Milford, MA, U.S.A.). Solvents and other chemicals of analytical grade were purchased from Beijing Chemical Engineering Company (Beijing, China).
Preparation of PMR and PMRPPMR and PMRP were purchased from Tong Ren Tang medicinal materials Co., Ltd., (Fig. 1). PMR and PMRP were accurately weighed, soaked for 1 h, and boiled for 1 h. After filtration, a 10-times volume of distilled water was added and boiled for 1 h again, then the mix was filtrated. The two filtrates were combined and rotary evaporated to 1g/mL.

(Color figure can be accessed in the online version.)
We performed UPLC/Q-TOF-MS to detect the chemical profiles of PMR and PMRP. The Chromatography conditions were as follows. Column was Waters HSS T3 (2.1 × 100 mm, 1.8 µm); mobile phase: A was an aqueous solution containing 0.1% formic acid, B was an acetonitrile solution containing 0.1% formic acid; flow rate was 0.5 mL/min; injection volume was 5 µL. Column temperature was 30°C. The mobile phase was eluted with gradient: 0–2 min, 20% B; 2–6 min, 20–30% B; 6–10 min, 30–50% B; 10–12 min, 50–80% B; 13–14 min, 80–100% B; 14–16 min, 20% B. DAD full wavelength scanning range was 200–400 nm.
Animal TreatmentFifteen male Sprague–Dawley rats (weighing 180–220 g each) were obtained from animal experimental center of Academy of Military Medical Science. Rats were housed with temperature and humidity at 22 ± 2°C and 45 ± 15%, under a natural light–dark cycle. All animal experiment protocols were carried out in accordance with the guidelines established by the animal experimental center of Academy of Military Medical Sciences (IACUC-AMMS-13-2017-012, May 2017). After 1 week acclimation period, rats were randomly divided into three groups, with five rats each, Control group, PMR group and PMRP group. Rats were all administered 1920 mg/kg/d orally for 28 d.28)
Sample PreparationAfter 28 d administrated, all rats were bled through the orbital iliac vein. After an allotted clotting time of the plasma solution of 10 min, all plasma samples were centrifuged at 3000 rpm for 10 min. Two hundred micro liters aliquot of plasma samples were added 800 µL of methanol, the resulting solution mixture was vortexed for 30 s and then centrifuged at 13000 rpm for 15 min at 4°C. All supernatant samples were transferred to new RNAase free tubes and evaporated to full dryness under stream of nitrogen. The residue was dissolved with 200 µL of methanol, followed by vortexing for 60 s and centrifuging at 13000 rpm for 15 min. The supernatant was transferred to a sampling vial for UPLC/Q-TOF-MS analysis.29) Quality control samples were prepared by pooling aliquots from all samples. Serum samples were allowed to thaw on ice at 4°C for 30 min, then a 100 µL aliquot of serum was added to a labeled 1.5 mL microcentrifuge tube, and 300 µL of acetonitrile was subsequently added. The preparation was thoroughly mixed on a vortex mixer for 15 s, then protein precipitate was pelleted by centrifugation at 12000 rpm for 10 min at 4°C. Finally, 100 µL of supernatant was transferred to a 200 µL vial for further analysis.
Biochemical Analysis and Histopathological ExaminationWhole blood was centrifuged at 3000 rpm for 15 min at 4°C to obtain the serum. levels of alanine aminotransferase (ALT), Aspartate aminotransferase (AST), alkaline phosphatase (ALP), lactate dehydrogenase (LDH), and total bilirubin (TBIL) were tested by using biochemical analyzer (Rayto, Shenzhen, China). Liver tissues were fixed in 10% neutral buffered formalin for 24 h, then embedded in paraffin, cut into sections (about 4–5 µm thick) by using a microtome. Hematoxylin–eosin (H&E) and immunohistochemistry (IHC) staining were used for highlighting the liver damage. For H&E staining, liver tissues were prepared and stained with H&E stain. For IHC Staining, the activity of endogenous peroxidase in the tissue sections or fixed cells was blocked with 3% hydrogen peroxide solution. The antigens were retrieved, and the nonspecific binding was blocked by 3% bovine serum albumin (BSA; Roche). Subsequently, tissue sections or cell coverslips were incubated with different primary antibodies (Anti-CD3 antibody (ab135372) (1 : 150) and Anti-CD4 antibody (ab183685) (1 : 200) (purchased from Abcam Shanghai, China), followed by incubation with horseradish peroxidase (HRP) conjugated goat anti-rabbit immunoglobulin G (IgG) (1 : 1000, Cell Signaling, MA, U.S.A.). Then, 3,3′-diaminobenzidine (DAB) chromogen substrate solution was utilized to visualize the results.30)
UPLC/Q-TOF-MS Analysis ConditionsChromatographic ConditionsHPLC-Q-TOF-MS system was used for serum metabolic spectrum analysis. One microliter aliquot of each sample was injected into the system on a HSS T3 column analytical column (2.1 × 100 mm, 1.8 µm, waters) for sample separation at 45°C. Solvent A (acetonitrile/water (60/40)) and solvent B (isopropanol/acetonitrile (90/10)) were used as the mobile phase for a linear gradient separation at a fow rate of 0.30 mL/min for 20 min with a linear gradient of 100% A over 0–1.0 min, 100–60% A over 1.0–9.0 min, 60–10% A over 9.0–19.0 min, 10–0% A over 19.0–20.0 min. Both positive and negative mode electrospray ionization sources were used. For hydrophilic interaction liquid chromatography (HILIC) separation, mobile phase A was acetonitrile and mobile phase B was water; both A and B contained 0.1% formic acid and 10 mmol/L ammonium acetate. The column was a BEH Amide column (2.1 × 100 mm, 1.7 µm, Waters) operated at 40°C. Column separation was performed by different gradient elution program. For 0 min, 80% A; 2 min, 70% A; 5 min 55% A; 6.5 min 40% A; 12 min 35% A; 14 min, 15% A; 17.5 min, 0% A; 18 min 0% A; 18.1 min 80% A.
MS ConditionsA Thermo Scientific™ Q Exactive™ hybrid quadrupole Orbitrap mass spectrometer equipped with a HESI-II probe was employed. Positive and negative HESI-II spray voltages were 3.7 and 3.5 kV, respectively. The heating capillary temperature was 320°C. Sheath gas pressure was 30 psi, auxiliary gas pressure was 10 psi, and collision gas pressure was 1.5 mTorr. Heated vaporizer temperature was 300°C. Sheath gas, auxiliary gas, and collision gas were all nitrogen. Parameters of the full mass scan were as follows: a resolution of 70000, an auto gain control target under 1 × 106, a maximum isolation time of 50 ms, and a m/z range 50–1500. LC-MS system was controlled by using Xcalibur 2.2 SP1.48 software (Thermo Fisher Scientific). Data were collected and processed.
Quality Control AnalysisWe took 20 µL from each prepared samples extract and mix to get the quality control (QC) samples. They were injected among every three detecting samples throughout the analytical run. We selected features based on samples’ coefficients of variation, sample features with coefficients of variation over 15% were eliminated.29)
Data Processing and Pattern Recognition AnalysisRaw MS data were analyzed using MarkerLynx Applications Manager (Version 4.1). Data were imported to SIMCA-P software (v14.0, Umetric, Umea, Sweden) for principal component analysis (PCA) and orthogonal to partial least-squares-discriminate analysis (OPLS-DA) after the mean-centered and pareto-scaled procedures. PCA mode was assessed by the intercepts of R2X and Q2, while OPLS-DA mode was assessed by the intercepts of R2Y and Q2 in permutation test to avoid overfitting. Metabolite identification was processed based on results, database, and standards verification. Potential biomarkers were selected by variable importance in the projection (VIP). Ions with VIP > 1 were considered as relevant metabolites to explain the classification. Human Metabolome Database (HMDB) was exploited to identify the potential biomarkers and further elucidate the mechanisms.26) For visual description of significant changes, heat map and correlation analysis of biomarkers to biomarkers and pathway enrichment were generated by MetaboAnalyst 4.0.
Statistical AnalysisANOVA and Duncan’s multi-range test were performed to investigate alterations of biochemical indicators. Data was all analysised by using SPSS 22.0 and presented with GraphPad Prism 7.0 (Graphpad Software, San Diego, CA, U.S.A.). Statistical significance was defined at p < 0.05.
UPLC/Q-TOF-MS analysis was applied to detect the chemical profiles of PMR and PMRP. The electrospray ionization (ESI) positive ion fingerprints were shown in Fig. 2. It can be seen that there is a significant difference in the chemical composition. The chemical profiles of Polygonum multiflorum Thunb have changed significantly before and after processing.

(Color figure can be accessed in the online version.)
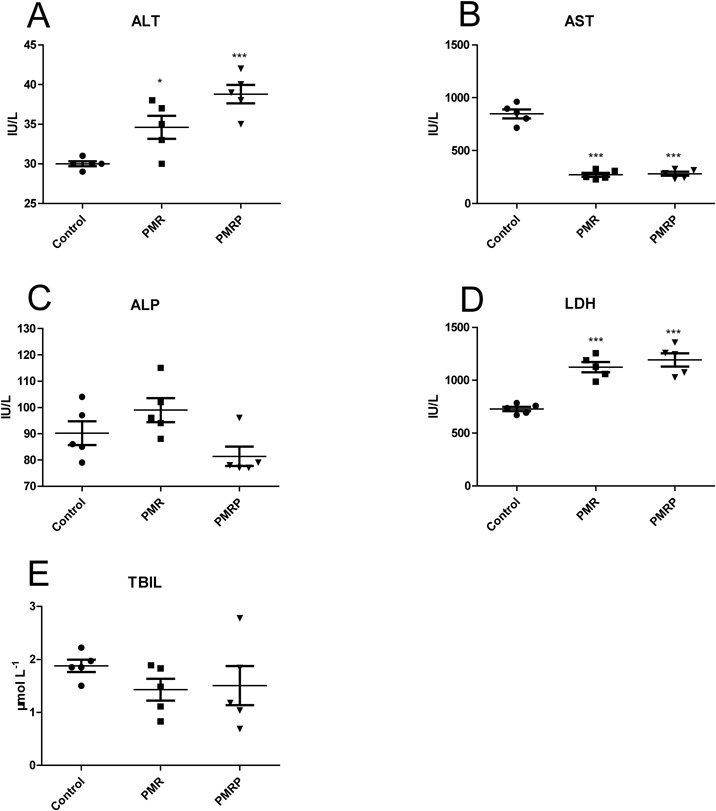
The levels of ALT and LDH were markedly increased in the PMR group and the PMRP group (Figs. 3A, D). Compared with the control group, the level of AST was markedly decreased in the PMR group and the PMRP group (Fig. 3B). The levels of ALP and TBIL had no obvious changes compared with the control group (Figs. 3C, E). serum parameters suggested that PMR and PMRP may have slight liver injury effects. H&E staining analysis showed the control group exhibited normal cellular structure (Fig. 4A), livers of the PMR group and the PMRP group presented no special changes (Figs. 4B, C). IHC results showed that there were almost no expression of CD3 and CD4 in control group (Figs. 4D, G). For CD3, there was slight expression in the RPMP group (Fig. 4E), and little higher expression in the PMR group (Fig. 4F). For CD4, there was no expression in the RPMP group (Fig. 4H), and slight expression in the PMR group (Fig. 4I). The results showed that PMR and PMR can cause mild immune inflammation.

For H&E staining, A. Control group, B. PMRP group, C. PMR group, Original magnification, ×400. For IHC staining; CD3 antibody: D. Control group; E. PMRP group; F. PMR group, CD4 antibody: G. Control group (saline); H. PMRP group; I. PMR group, original magnification, ×400. (Color figure can be accessed in the online version.)
QC can determine whether the systematic error of the whole experiment is within the controllable range.31) In this study, QC samples and experimental samples were analyzed using unsupervised PCA. PCA scores of ESI positive ion mode, negative ion mode and HILIC mode are shown (Figs. 5A–C). The cluster analysis of QC samples showed that they were closely clustered in three modes respectively. The results proved that the detecting method has good stability and repeatability.

A. positive ion mode, B. negative ion mode, C. HILIC mode. (Color figure can be accessed in the online version.)
PCA and OPLS-DA can represent the differences in metabolomic profiles. PCA can provide an overview of the information hidden in multivariate data, whereas OPLS-DA can enhance the separation among groups of observations and improve interpretation. In this study, PCA was employed to reveal the metabolic changes.There were clear differences between the PMR and PMRP groups (Fig. 6). PCA score plot showed a clear trend of group clustering. Three models’ parameter showed R2X of 0.942 and Q2 of 0.820, R2X of 0.917 and Q2 of 0.872, and R2X of 0.781 and Q2 of 0.640, respectively (Figs. 6A, 7A, 8A).

A. PCA score plot, B. OPLS-DA score plot, C. permutation test of OPLS-DA model, and D. S-plot of the OPLS-DA. (Color figure can be accessed in the online version.)

A. PCA score plot, B. OPLS-DA score plot, C. permutation test of OPLS-DA model, and D. S-plot of the OPLS-DA. (Color figure can be accessed in the online version.)

A. PCA score plot, B. OPLS-DA score plot, C. permutation test of OPLS-DA model, and D. S-plot of the OPLS-DA. (Color figure can be accessed in the online version.)
To capture the distinctive metabolic phenotypes and maximize the discrimination between PMR and PMRP groups by means of all the identified metabolites, we applied a supervised OPLS-DA model. The OPLS-DA score plots model indicated that the cluster of PMR group was well separated from PMRP group. The variation values of the OPLS-DA model were R2Y of 0.997 and Q2 of 0.982, R2Y of 0.997 and Q2 of 0.996, R2Y of 0.997 and Q2 of 0.987, respectively (Figs. 6B, 7B, 8B). These results indicate that the model was valid and reliable. Permutation plots figure displayed Q2 points that intersected the vertical axis below zero suggesting a low risk of overfitting, indicating that the original models were valid (Figs. 6C, 7C, 8C). Difference variables were also revealed by S-plot of the OPLS-DA models (Figs. 6D, 7D, 8D).
The parameter indicated that these metabolites had high sensitivity and specificity for the identification, therefore the metabolites could be used as metabolic biomarkers between the PMR group and PMRP group.
Identifcation of Potential MetabolitesTotal ion current diagrams are shown in Fig. 9. VIP values indicate the separating power between groups. VIP values of all the metabolites from the OPLS-DA model were confirmed. Metabolites which had a VIP value >1 were considered as biomarkers. The biomarkers are listed in Table 1. Fifty-two serum metabolites were identified as biomarkers. In positive ion mode, 31 biomarkers were identified, including PC (16 : 0/14 : 0), PC (16 : 0/16 : 0), Ceramide (d18 : 1/16 : 0), LysoPC (18 : 0), LysoPC (22 : 0), LysoPC (24 : 0), PE (18 : 2(9Z,12Z)/16 : 0), and PE (20 : 4(8Z,11Z,14Z,17Z)/16 : 0), etc. In negative ion mode, 13 biomarkers were identified, including glycodeoxycholic acid, chenodeoxycholic acid, lithocholic acid, (Z)-9-Heptadecenoic acid, β-muricholic acid, taurocholic acid, cholic acid, and glycochenodeoxycholic acid, etc. In HILIC mode, nine biomarkers were identified, including niacinamide, L-palmitoylcarnitine, butyrylcarnitine, L-acetylcarnitine, L-kynurenine, taurine, L-alanine, 3-methylhistidine, and ornithine.

A. PMR group with positive ion mode, B. PMRP group with positive ion mode, C. PMR group with negative ion mode, D. PMRP group with positive ion mode, E. PMR group with HILIC mode, F. PMRP group with HILIC mode. (Color figure can be accessed in the online version.)
| Mode | RTa) | VIPb) | FCc) | m/z | Formula | Identifier | CNd) | HMDB | KEGG |
|---|---|---|---|---|---|---|---|---|---|
| C18 + | 10.12 | 1.110 | 0.664 | 804.552 | C46H78NO8P | PC(18:2(9Z,12Z)/20:5(5Z,8Z,11Z,14Z,17Z)) | 1 | HMDB08149 | C00157 |
| 10.22 | 1.028 | 0.698 | 754.537 | C42H76NO8P | PC(20:4(8Z,11Z,14Z,17Z)/14:0) | 2 | HMDB08459 | C00157 | |
| 10.38 | 1.074 | 0.663 | 730.537 | C40H76NO8P | PC(14:0/18:2(9Z,12Z)) | 3 | HMDB07874 | C00157 | |
| 10.41 | 1.326 | 0.548 | 830.568 | C48H80NO8P | PC(20:4(8Z,11Z,14Z,17Z)/20:4(8Z,11Z,14Z,17Z)) | 4 | HMDB08477 | C00157 | |
| 10.59 | 1.166 | 0.611 | 756.552 | C42H78NO8P | PC(16:0/18:3(9Z,12Z,15Z)) | 5 | HMDB07975 | C00157 | |
| 10.60 | 1.265 | 0.565 | 780.552 | C44H78NO8P | PC(20:4(8Z,11Z,14Z,17Z)/16:1(9Z)) | 6 | HMDB08463 | C00157 | |
| 10.77 | 1.139 | 0.620 | 768.552 | C43H78NO8P | PC(20:4(5Z,8Z,11Z,14Z)/15:0) | 7 | HMDB08428 | C00157 | |
| 10.94 | 1.297 | 0.539 | 744.552 | C41H78NO8P | PC(18:2(9Z,12Z)/15:0) | 8 | HMDB08132 | C00157 | |
| 10.01 | 1.521 | 0.436 | 756.552 | C42H78NO8P | PC(18:3(6Z,9Z,12Z)/16:0) | 9 | HMDB08166 | C00157 | |
| 11.08 | 1.723 | 0.419 | 706.537 | C38H76NO8P | PC(16:0/14:0) | 10 | HMDB07965 | C00157 | |
| 11.70 | 1.056 | 0.642 | 740.521 | C41H74NO8P | PE(20:4(8Z,11Z,14Z,17Z)/16:0) | 11 | HMDB09418 | C00350 | |
| 11.90 | 1.464 | 0.452 | 716.522 | C39H74NO8P | PE(18:2(9Z,12Z)/16:0) | 12 | HMDB09088 | C00350 | |
| 11.91 | 1.060 | 0.671 | 746.568 | C41H80NO8P | PC(18:1(11Z)/15:0) | 13 | HMDB08066 | C00157 | |
| 12.08 | 1.293 | 0.543 | 796.584 | C45H82NO8P | PC(15:0/22:4(7Z,10Z,13Z,16Z)) | 14 | HMDB07955 | C00157 | |
| 12.39 | 1.165 | 0.646 | 834.598 | C48H84NO8P | PC(22:6(4Z,7Z,10Z,13Z,16Z,19Z)/18:0) | 15 | HMDB08727 | C00157 | |
| 12.43 | 1.352 | 0.551 | 734.568 | C40H80NO8P | PC(16:0/16:0) | 16 | HMDB00564 | C00157 | |
| 12.66 | 1.095 | 0.661 | 760.583 | C42H82NO8P | PC(16:0/18:1(11Z)) | 17 | HMDB07971 | C00157 | |
| 12.77 | 1.245 | 0.576 | 810.598 | C46H84NO8P | PC(20:4(8Z,11Z,14Z,17Z)/18:0) | 18 | HMDB08464 | C00157 | |
| 12.85 | 1.222 | 0.594 | 836.614 | C48H86NO8P | PC(18:0/22:5(4Z,7Z,10Z,13Z,16Z)) | 19 | HMDB08055 | C00157 | |
| 13.01 | 1.390 | 0.502 | 786.599 | C44H84NO8P | PC(18:0/18:2(9Z,12Z)) | 20 | HMDB08039 | C00157 | |
| 13.04 | 1.120 | 1.804 | 538.518 | C34H67NO3 | Ceramide (d18:1/16:0) | 21 | HMDB04949 | C00195 | |
| 13.23 | 1.287 | 0.538 | 768.554 | C43H78NO8P | PC(20:4(8Z,11Z,14Z,17Z)/15:0) | 22 | HMDB08461 | C00157 | |
| 14.17 | 1.224 | 0.614 | 762.600 | C42H84NO8P | PC(18:0/16:0) | 23 | HMDB08034 | C00157 | |
| 14.36 | 1.100 | 0.659 | 788.615 | C44H86NO8P | PC(18:0/18:1(11Z)) | 24 | HMDB08037 | C00157 | |
| 14.59 | 1.281 | 0.570 | 814.631 | C46H88NO8P | PC(18:0/20:2(11Z,14Z)) | 26 | HMDB08045 | C00157 | |
| 2.64 | 1.069 | 0.655 | 518.323 | C26H48NO7P | LysoPC(18:3(6Z,9Z,12Z)) | 26 | HMDB10387 | C04230 | |
| 5.86 | 1.049 | 0.667 | 524.370 | C26H54NO7P | LysoPC(18:0) | 27 | HMDB10384 | C04230 | |
| 6.17 | 1.001 | 0.691 | 550.386 | C28H56NO7P | LysoPC(20:1(11Z)) | 28 | HMDB10391 | C04230 | |
| 8.85 | 1.130 | 0.633 | 580.433 | C30H62NO7P | LysoPC(22:0) | 29 | HMDB10398 | C04230 | |
| 8.88 | 1.160 | 0.623 | 606.449 | C32H64NO7P | LysoPC(24:1(15Z)) | 30 | HMDB10406 | C04230 | |
| 9.97 | 1.137 | 0.633 | 608.464 | C32H66NO7P | LysoPC(24:0) | 31 | HMDB10405 | C04230 | |
| C18− | 10.10 | 1.642 | 0.228 | 448.307 | C26H43NO5 | Glycodeoxycholic acid | 32 | HMDB0000631 | C05464 |
| 11.24 | 1.604 | 0.235 | 391.286 | C24H40O4 | Chenodeoxycholic acid | 33 | HMDB0000518 | C02528 | |
| 12.85 | 1.487 | 0.253 | 375.290 | C24H40O3 | Lithocholic acid | 34 | HMDB0000761 | C03990 | |
| 13.27 | 1.079 | 2.375 | 227.201 | C14H28O2 | Myristic acid | 35 | HMDB00806 | C06424 | |
| 14.62 | 1.095 | 2.455 | 267.233 | C17H32O2 | (Z)-9-Heptadecenoic acid | 36 | HMDB31046 | C16536 | |
| 16.56 | 1.085 | 2.457 | 297.280 | C19H38O2 | Nonadecanoic acid | 37 | HMDB00772 | C16535 | |
| 7.15 | 1.366 | 0.309 | 407.280 | C24H40O5 | β-Muricholic acid | 38 | HMDB0000415 | C17726 | |
| 8.23 | 1.700 | 0.283 | 514.284 | C26H45NO7S | Taurocholic acid | 39 | HMDB0000036 | C05122 | |
| 8.34 | 2.051 | 0.084 | 464.301 | C26H43NO6 | Glycocholic acid | 40 | HMDB0000138 | C01921 | |
| 9.09 | 1.437 | 0.279 | 391.285 | C24H40O4 | Ursodeoxycholic acid | 41 | HMDB0000946 | C07880 | |
| 9.60 | 1.551 | 0.219 | 407.280 | C24H40O5 | Cholic acid | 42 | HMDB0000619 | C00695 | |
| 9.75 | 2.450 | 0.026 | 448.307 | C26H43NO5 | Glycochenodeoxycholic acid | 43 | HMDB0000637 | C05466 | |
| Hilic | 1.33 | 1.265 | 0.659 | 123.055 | C6H6N2O | Niacinamide | 44 | HMDB01406 | C00153 |
| 1.38 | 1.023 | 0.763 | 400.341 | C23H45NO4 | L-Palmitoylcarnitine | 45 | HMDB00222 | C02990 | |
| 2.19 | 1.718 | 0.483 | 232.154 | C11H21NO4 | Butyrylcarnitine | 46 | HMDB02013 | C02862 | |
| 3.16 | 1.012 | 0.768 | 204.123 | C9H17NO4 | L-Acetylcarnitine | 47 | HMDB00201 | C02571 | |
| 4.78 | 1.461 | 0.580 | 209.092 | C10H12N2O3 | L-Kynurenine | 48 | HMDB00684 | C00328 | |
| 5.31 | 1.686 | 0.567 | 126.022 | C2H7NO3S | Taurine | 49 | HMDB00251 | C00245 | |
| 5.86 | 1.340 | 0.626 | 90.055 | C3H7NO2 | L-Alanine | 50 | HMDB00161 | C00041 | |
| 7.05 | 1.664 | 0.486 | 170.092 | C7H11N3O2 | 3-Methylhistidine | 51 | HMDB00479 | C01152 | |
| 7.35 | 1.016 | 1.339 | 133.097 | C5H12N2O2 | Ornithine | 52 | HMDB00214 | C00077 |
a) RT, retention time, b) VIP, Variable Importance in the Projection. c) FC, fold change. d) CN, Corresponding number.
In order to investigate whether differences in metabolic pathways between the PMR and PMRP groups, the metabolic pathways were constructed by importing the identifed potential metabolites’ HMDB into MetaboAnalyst 4.0. It is a useful online tool for analyzing complex and intertwining or enriching metabolic pathways.32,33) The p-value of pathway impact was calculated. The threshold was set to 0.01, and values above this threshold were filtered out as significant pathways. HMDB of biomarkers were imported into the MetaboAnalyst 4.0 to find the specific pathways.
A clustering heat map directly revealed the variation of biomarkers (Fig. 10). Fold change is often used to further investigate the magnitude of change of biomarkers. Compared with the PMR group, the PMRP group showed increased levels of ceramide (d18 : 1/16 : 0), myristic acid, (Z)-9-heptadecenoic acid, and nonadecanoic acid, whereas the levels of other biomarkers were decreased (Table 1). Metabolite correlation analysis was revealed by heat map plotting (Fig. 11). We performed the metabolites sets enrichment analysis. The top six routes for scoring were taurine and hypotaurine metabolism, sphingolipid metabolism, glycerophospholipid metabolism, nicotinate and nicotinamide metabolism, arginine and proline metabolism, and primary bile acid biosynthesis. Bile acid biosynthesis was the most differential pathway (Figs. 12A, B, Table 2). Clicking the corresponding node (Fig. 13A), we can obverve the pathway view on the panel (Fig. 13). Labels within small boxes correspond to KEGG identifiers for metabolites. In Fig. 13A, the metabolites were taurine (C00245, HMDB0000251) and taurocholic acid (C05122, HMDB0000036). In Fig. 13B the metabolite was N-acylsphingosine (C00195, HMDB0004947). In Fig. 13C, the metabolites were phosphatidylcholine (C00157, HMDB00564), phosphatidylethanolamine (C00350, HMDB05779) and lyso-phosphatidylcholine (lysoPC) (18 : 1(9Z)) (C04230, HMDB0002815). In Fig. 13D, the metabolites were phosphatidylcholine (C00157, HMDB00564) and phosphatidylethanolamine (C00350, HMDB05779). In Fig. 13E, the metabolite was L-ornithine (C00077, HMDB0000214). In Fig. 13F, the metabolites were taurine (C00245, HMDB0000251), cholic acid (C00695, HMDB0000619), glycocholic acid (C01921, HMDB0000138), taurocholic acid (C05122, HMDB0000036), glycochenodeoxycholic acid (C05466, HMDB0000637), and chenodeoxycholic acid (C02528, HMDB0000518).
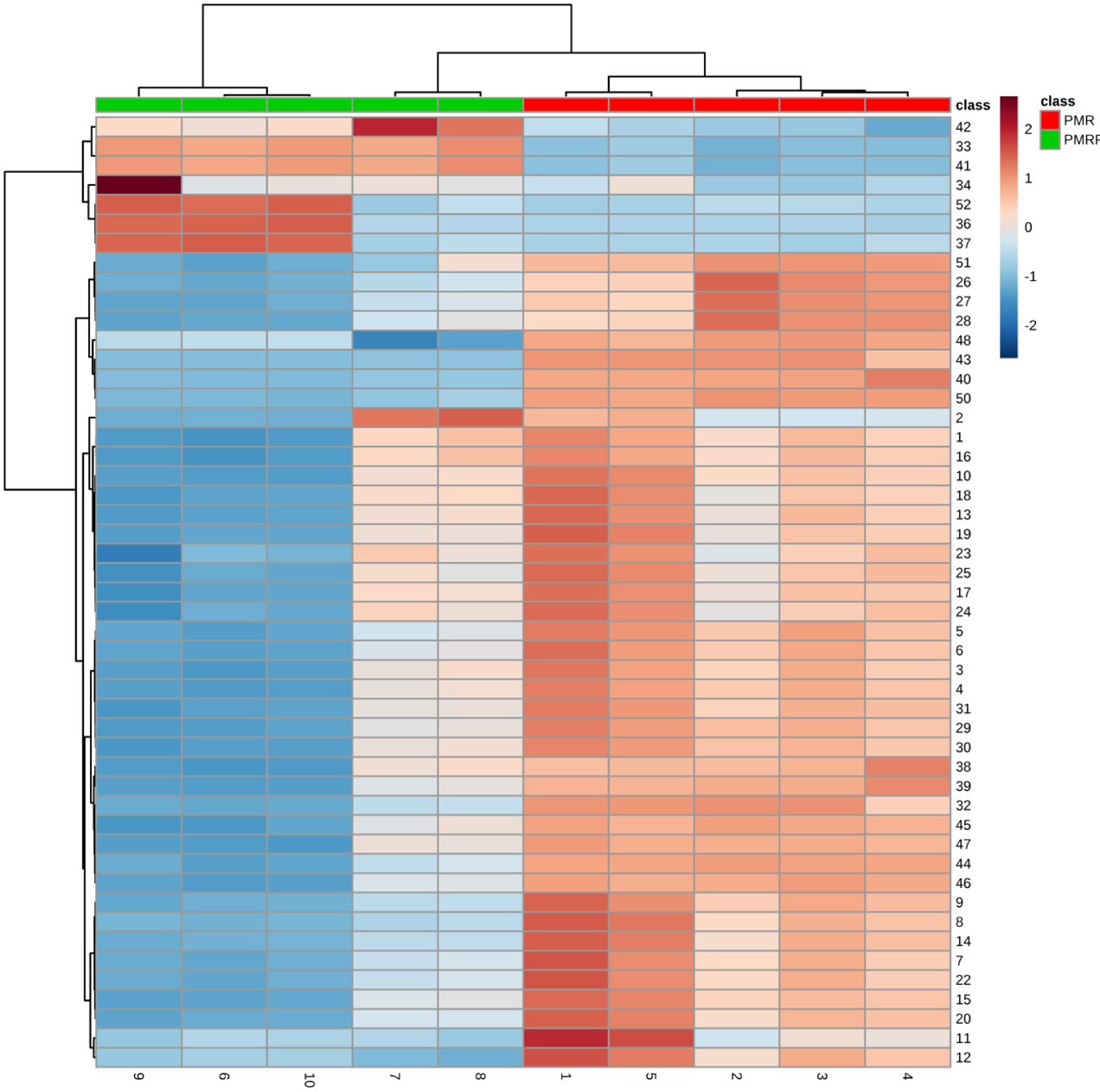
Rows: samples; Columns: biomarkers (Numbers 1–10 below represent samples in each group). (Color figure can be accessed in the online version.)

(Color figure can be accessed in the online version.)

(Color figure can be accessed in the online version.)

A. Pathway of taurine and hypotaurine metabolism B. Pathway of sphingolipid metabolism C. Pathway of glycerophospholipid metabolism D. Pathway of nicotinate and nicotinamide metabolism E. Pathway of arginine and proline metabolism F. Pathway of primary bile acid biosynthesis. (Color figure can be accessed in the online version.)
| Main pathway | Totala) | Hitsb) | Raw Pc) | Holm Pd) | −log(P)e) | Impactf) |
|---|---|---|---|---|---|---|
| Primary bile acid biosynthesis | 46 | 6 | 0.0001 | 0.0077 | 9.279 | 0.1190 |
| Taurine and hypotaurine metabolism | 8 | 2 | 0.0078 | 0.6354 | 4.848 | 0.4286 |
| Glycerophospholipid metabolism | 30 | 3 | 0.0144 | 1 | 4.241 | 0.2750 |
| Nicotinate and nicotinamide metabolism | 13 | 1 | 0.2074 | 1 | 1.573 | 0.2381 |
| Sphingolipid metabolism | 21 | 1 | 0.3137 | 1 | 1.159 | 0.2807 |
| Arginine and proline metabolism | 44 | 1 | 0.5486 | 1 | 0.600 | 0.1274 |
a) Total: the total number of compounds in the pathway. b) Hits: the matched number of metabolites in one pathway. c) Raw P: the original p value calculated from the enrichment analysis. d) Holm P: the p value further adjusted using Holm–Bonferroni method. e) −log(P): Y-axis values. f) Impact: the pathway impact value calculated from pathway topology analysis.
Herbal medicine has become the second most common cause of drug-induced liver damage (DILI) in the United States.34) Rare and unpredictable herbal drug induced liver damage (HILI) with idiosyncratic reactions is well documented.35,36) Clinical features of liver injury by TCM drugs are variable, but specific diagnostic biomarkers are available for only a small subset of herbs, and reliable methods to identify the DILI of herbs remain elusive17,37) despite normal metabolic function of liver being compromised.38) Take PMR for example, evidence chain-based causality identification in HILI has been successfully studied.34) PMR has a wide range of clinical applications, but clinical DILI reports involving PMR are increasing.39) Due to the special nature of TCM drugs and complexity of clinical use, it is difficult to identify and evaluate the toxicity.40) Concurrently, the direct or indirect long-term DILI effects of TCM drugs are difficult to determine.
Recent research elucidating the mechanism of liver injury has focused on metabolomic methods in chronic liver injury caused by PMR or PMRP using specific animal models.41) Research has shown that toxicity of PMR is not dependent on the content of anthranoid derivatives but may correlate with the content of tetrahydroxystilbene glucosides.42) PMR did not cause acute toxicity. The toxic effects occurred when patients had taken a large dose of PMR over a prolonged period. A logical assumption being that the accumulation of toxic components in vivo resulted in the presentation of these toxic events over time.43,44) Furthermore, the acute toxicity caused by PMR typically occurred in patients with idiopathic symptoms. Relevant ingredients in PMR had a direct link to specific liver damage.34,41,45) Ruling out patients with idiosyncrasies, it is difficult to find chronic liver damage caused by PMR for ordinary patients. Researchers found that PMRP has a dose-time-toxicity dependent correlation. Serum metabonomics studies revealed 12 characteristic metabolites were obtained through metabolomic analysis and 7 metabolic pathways involved, resulting in significant disturbance in amino acid metabolism, energy metabolism, and bile acid metabolism.14)
Changes in endogenous metabolic profiles are more sensitive and easier to observe. Here, results show that bile acid metabolism is one of the most metabolic pathways in rats between PMR group and PMRP group. Recent research has indicated that anthraquinones derived from PMR can alter bile acid disposition through direct inhibition of bile acid transporters as well as regulated expression of bile acid transporters and enzymes.46) Due to the slow onset of a pharmacologically relevant effect, TCM herbs are typically taken over a long period. It is difficult to observe liver damage using biochemical indicators.
Metabolomics provide an opportunity to develop the systematic analysis of metabolites and have been successfully applied to establishing biomarkers and perturbed pathways which, in turn, can clarify the mechanism of action of traditional Chinese medicine.47–49) In this study, results show that biochemical indicators (such as ALT, AST, ALP, LDH, and TBIL) cannot be used as indicators of liver damage caused by PMR and PMRP. A nontargeted metabolic profile based on UPLC/Q-TOF-MS was developed to investigate significant metabolic changes in rat serum between PMR and PMRP groups. A holistic metabolic profile of serum was detected. Six pathways (taurine and hypotaurine metabolism, sphingolipid metabolism, glycerophospholipid metabolism, nicotinate and nicotinamide metabolism, arginine and proline metabolism, and primary bile acid biosynthesis) were confirmed as the most significant pathways.
Taurine exists in a free state in the body, it participates in protein synthesis. It has antioxidant and osmotic pressure regulation, and regulates bile acid binding, ion movement, and nerve transmission. In liver, the role of taurine is to combine with bile acids to form taurocholic acid, which is essential for the absorption of lipids in the digestive tract.50–53) Taurine and hypotaurine metabolism play an important role in bile acid transportation. Sphingolipids are important components of biofilm structure. Sphingolipids and their metabolites are important active molecules involved in many important signal transduction processes such as cell growth, differentiation, senescence, and programmed cell death.54,55) Glycerol phospholipid is the most abundant phospholipid in the body. It is an important component of bile and membrane surfactants can be involved in the recognition and signal transduction of proteins by cell membranes.56) Studies have indicated that PMR might have a more satisfactory effect in the clinical treatment of nonalcoholic fatty liver disease (NAFLD) or hyperlipidemia characterized by the elevation of cholesterol than processed PMR.26) Nicotinate (vitamin B3) is one of the 13 essential vitamins in the body. Nicotinate can convert into nicotinamide. Niacinamide is a component of coenzymes I and II, which participate in lipid metabolism, oxidation processes of tissue respiration, and anaerobic decomposition of carbohydrates.57,58) Arginine and proline participate in the ornithine cycle, promote the formation of urea, and convert the ammonia produced into a non-toxic urea through the ornithine cycle, which is discharged in the urine, thereby reducing the blood ammonia concentration.59) The arginine and proline metabolism pathway is affected by PMR and PMRP in this study.
Bile acids play an important role in lipid metabolism. Bile acids are mainly found in the enterohepatic circulatory system and play a protective role through recycling. Only small fraction of bile acids enter into the peripheral circulation. Though bile acids have been known as digestive juice, recent studies have revealed that bile acids act as signaling molecules to control metabolism and inflammation.60–62) Bile acids and urine tauro-beta-muricholic acid were considered promising biomarkers among PMR induced hepatotoxicity.
In this study, a metabolomic profile based on UPLC/Q-TOF-MS analysis was developed to investigate changes in the rat serum among water extracts between PMR and PMRP. Fifty-two biomarkers were confirmed, and six metabolic pathways, including taurine and hypotaurine metabolism, sphingolipid metabolism, glycerophospholipid metabolism, nicotinate and nicotinamide metabolism, arginine and proline metabolism, and tryptophan metabolism and primary bile synthesis were confirmed by MetaboAnalyst 4.0. This is first metabolomic study to identify potential differential endogenous metabolites and pathways that may be related to water extracts of between PMR and PMRP. However, the limitation is that we do not confirm the mechanisms. We will explore the effects of metabolite identification on pathway exploration in order to find out the hepatotoxicity mechanisms caused by PMR and PMRP. We will be focusing on identifying the specific compounds that may contribute to hepatotoxicity.
This work was supported by the National Major Science and Technology Project (No. 2015ZX09501004-003-003).
The authors declare no conflict of interest.