2022 Volume 45 Issue 10 Pages 1559-1563
2022 Volume 45 Issue 10 Pages 1559-1563
Dihydroceramide Δ4-desaturase 1 (DEGS1) enzymatic activity is inhibited with N-(4-hydroxyphenyl)-retinamide (4-HPR). We reported previously that 4-HPR suppresses severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) entry through a DEGS1-independent mechanism. However, it remains unclear whether DEGS1 is involved in other SARS-CoV-2 infection processes, such as virus replication and release. Here we established DEGS1 knockout (KO) in VeroE6TMPRSS2 cells. No significant difference was observed in virus production in the culture supernatant between wild-type (WT) cells and DEGS1-KO cells, although the levels of dihydroceramide (DHCer), a DEGS1 substrate, were significantly higher in DEGS1-KO cells than WT cells. Furthermore, the virus-induced cytopathic effect was also observed in DEGS1-KO cells. Importantly, the EC50 value of 4-HPR in DEGS1-KO cells was almost identical to the value reported previously in WT cells. Our results indicated the lack of involvement of DEGS1 in SARS-CoV-2 infection.
The coronavirus disease (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), originated from Wuhan, Hubei Province, China.1–4) By the end of May 2022, more than 450 million people have been infected worldwide, with more than 6.3 million deaths have been documented in 225 countries (https://www.worldometers.info/coronavirus/). The development of vaccines against SARS-CoV-2 has significantly reduced the spread of the virus. In addition, remdesivir, an inhibitor of viral RNA polymerase, has received an emergency use authorization for the treatment of COVID-19 because it shortened the recovery time of patients treated in clinical trials.5) The development of new inhibitors of SARS-COV-2 is needed to enhance clinical efficacy and increase the options for combination therapy for COVID-19.
We investigated previously the roles of sphingolipids in SARS-CoV-2 spike protein (S)-meditated membrane fusion by the cell–cell fusion assay using SARS-CoV-2 mimicked HEK293FT cells and host mimicked HEK293FT cells treated with an inhibitor of the enzyme involved in sphingolipid metabolism.6) N-(4-Hydroxyphenyl)-retinamide (4-HPR), an inhibitor of dihydroceramide Δ4-desaturase 1 (DEGS1), potently suppressed SARS-CoV-2 S-mediated membrane in a cell–cell fusion assay system (50% effective concentration: EC50 = 4.1 µM). Furthermore, 4-HPR exhibited potent antiviral activity against SARS-CoV-2 with EC50 value of 4.4 µM in VeroE6TMPRSS2 cells. Since the efficiency of cell–cell fusion in DEGS1-KO HEK293T cells was at a level comparable to that in wild-type (WT) cells, we concluded that 4-HPR suppresses SARS-CoV-2 S-mediated membrane fusion through a DEGS1-independent mechanism. However, it remains unclear whether DEGS1 is involved in other life cycles, such as virus replication and release. During the replication process and the release of viral progeny, a cytopathic effect occurs that damages the host cell and ultimately results in cell death.
The purpose of this study was to determine the role of DEGS1 in SARS-CoV-2 infection. For this purpose, we investigated DEGS1 function in the entire process of SARS-CoV-2 infection, rather than the entry stage only, using VeroE6TMPRSS2 cells that exhibit the cytopathic effects of the virus. We created DEGS1-KO VeroE6TMPRSS2 cells using the clustered regularly interspaced short palindromic repeats CRISPR-associated proteins 9 (CRISPR/Cas9) gene-editing system, and analyzed the function of DEGS1 in virus infection with the SARS-CoV-2 cytopathic effect assay.
VeroE6TMPRSS2 cells were obtained from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan).7) The cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS, 100 µg/mL of penicillin, 100 µg/mL of streptomycin, and 1 mg/mL of geneticin. SARS-CoV-2 NCGM-05-2N strain (SARS-CoV-205-2N) was isolated from nasopharyngeal swabs of a patient with COVID-19, who had been admitted to the National Center for Global Health and Medicine, Tokyo, Japan.
Chemicals and Antibodies4-HPR was obtained from Tokyo Chemical Industry (Tokyo, Japan). The compound was dissolved in dimethyl sulfoxide (DMSO) and diluted to a final concentration of 0.2% DMSO in cell culture medium. N-palmitoyl-d31-D-erythro-sphingosine (D31-Cer) was obtained from Avanti Polar Lipids (Alabaster, AL, U.S.A.).
Rabbit polyclonal anti- ACE2 antibody (catalog #ab15348), rabbit immunoglobulin G (IgG) monoclonal anti-TMPRSS2 antibody (catalog #ab92323), rabbit IgG monoclonal anti-DEGS1 antibody (catalog #ab167169) were obtained from Abcam (Cambridge, U.K.). Goat horseradish peroxidase-conjugated anti-rabbit IgG antibody (catalog #7074S) was obtained from Cell Signaling Technology (Beverly, MA, U.S.A.). Mouse IgG monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (catalog #014-25524) was obtained from FUJIFILM (Tokyo). Mouse IgG1 monoclonal anti-ACE2 antibody (catalog #66699-1-Ig) was obtained from ProteinTech (Rosemont, IL, U.S.A.). Mouse IgG1 isotype control was obtained from BioLegend (San Diego, CA, U.S.A.). Goat PE-conjugated anti-mouse IgG polyclonal antibody (catalog #12-4010-82) was obtained from Thermo Fisher Scientific (Waltham, MA, U.S.A.).
Generation of CRISPR/Cas9-Based DEGS1-KO CellsTo establish DEGS1-KO cells, we designed guide RNAs (pSpCas9-DEGS1 gRNA-For, CACCGATGGATCATCATCGGTACCT; pSpCas9-DEGS1 gRNA-Rev, AAACAGGTACCGATGATGATCCATC) using the target design software (http://crispor.tefor.net/). The corresponding sequences to these sgRNAs were cloned into the pSpCas9(BB)-2A-GFP (PX458) vector, which was a kind gift from Feng Zhang (Addgene plasmid #48138).8) The constructs were transfected into VeroE6TMPRSS2 cells using Lipofectamine 2000 (Invitrogen MA, U.S.A.) according to the instructions supplied by the manufacturer. The transfected cells were cultured for 48 h and selected by the GFP marker using FACS Aria II (BD Biosciences, San Jose, CA, U.S.A.). Clonal populations of DEGS1-KO cells were isolated using limiting dilution. Disruption of DEGS1 was confirmed by DNA sequencing and quantitative sphingolipid metabolome of the cells using LC-tandem mass spectrometry (LC-MS/MS).
Analysis of Cell-Surface Expression of ACE2The details of this procedure have been described previously.6) Cells were fixed with 4% formaldehyde before incubation with either anti-ACE2 antibody (ProteinTech) or mouse IgG1 isotype control (BioLegend) antibody at 4 °C. Cells were stained with PE-conjugated secondary antibody (Thermo Fisher Scientific) and analyzed using the FACSCanto II instrument (BD Biosciences). Data were analyzed using the FlowJo software (Tree Star Inc., San Carlos, CA, U.S.A.).
Lipid Extraction and Quantification of Sphingolipids by LC-MS/MSThe details of this procedure have been described previously.6) Cells were washed once with cold PBS, collected in 100 µL of cold PBS, and then homogenized using sonication. Part of the sample (5 µL) was used in the bicinchoninic acid protein assay to determine the amount of protein. Total lipids were extracted by adding 375 µL of chloroform: methanol (1 : 2, v/v) containing 40 pmol of D31-Cer as internal standards. After sonicating the single-phase mixture, 100 µL of chloroform: methanol: 5N NaOH (1 : 2 : 0.8, v/v/v) was added and the solution was incubated for 1 h at 37 °C, followed by neutralization with acetic acid. Subsequently, 158 µL of chloroform and 158 µL of water were added and the mixture vigorously vortexed, before centrifuging for 1 min at 13000 × g at 4 °C. The lower phase was withdrawn and dried and then resuspended in acetonitrile: methanol (1 : 1, v/v), sonicated for 10 s, centrifuged at 14000 × g for 5 min, and the supernatant was transferred to vials. The concentrations of Cer and DHCer were analyzed using the QTRAP4500 instrument (SCIEX, Framingham, MA, U.S.A.).
SARS-CoV-2 Infectivity AssayVeroE6TMPRSS2 and DEGS1-KO VeroE6TMPRSS2 cells were seeded onto 96-well plates (1 × 104 cells/well). On the following day, the cells were inoculated with SARS-CoV-205-2N at a multiplicity of infection (MOI) of 0.01. To evaluate the infectivity of SARS-CoV-205-2N, the cell culture supernatant was harvested at days 0, 1, 2, 3 and viral RNA was extracted using QIAamp Viral RNA Mini Kit (QIAGEN, Germany). Then quantitative RT-PCR (RT-qPCR) was performed using One Step PrimeScript III RT-qPCR Mix (TaKaRa Bio, Japan) according to the protocol provided by the manufacturer. The primers and probe used for detection of SARS-CoV-2 nucleocapsid were: 5′-AAATTTTGGGGACCAGGAAC-3′ (forward), 5′-TGGCAGCTGTGTAGGTCAAC-3′ (reverse), and 5′-FAM-ATGTCGCGCATTGGCATGGA-BHQ-3′ (probe) (PMID: 32074516, 32165541).
SARS-CoV-2 Cytopathic Effect AssayDEGS1-KO VeroE6TMPRSS2 cells were seeded onto 96-well plates (5 × 103 cells/well). On the following day, the cells were cultured with 4-HPR for 3 d before inoculation of SARS-CoV-205-2N at a MOI of 0.01. After the 3-d culture, the level of cytopathic effect was determined in SARS-CoV-2-exposed cells using the WST-8 assay employing Cell Counting Kit-8 (Dojindo, Kumamoto, Japan).
Statistical AnalysisAll data were expressed as mean ± standard deviation (S.D.). The Student t-test was used for comparisons of two groups. A p-value of <0.05 was set to denote statistically significant difference. All statistical analyses were performed with GraphPad Prism 6 (GraphPad Software, San Diego, CA, U.S.A.).
First, we established DEGS1-KO VeroE6TMPRSS2 cells using the CRISPR/Cas9 gene-editing system. Sequence analysis showed that the 5-bp deletion of DEGS1 within exon 2 caused frame-shift mutation (Fig. 1A). The loss of DEGS1 was examined by Western blot analysis. As shown in Fig. 1B, immunoblotting showed the loss of DEGS1 expression in the DEGS1-KO cells. Importantly, comparable expression levels of ACE2 and TMPRSS2 (receptor and cofactor for virus entry, respectively) were noted in DEGS1-KO cells and WT cells. Similar results were obtained in FACS analysis of cell-surface ACE2 expression, although an appropriate antibody to detect the cell-surface TMPRSS2 was unavailable in this study. No significant differences were observed in the expression levels of ACE2 on the cell surface between WT cells and DEGS1-KO cells (Fig. 1C). These results indicate the protocol used to prepare DEGS1-KO cells resulted in complete loss of DEGS1 expression without affecting ACE2 and TMPRSS2 levels.
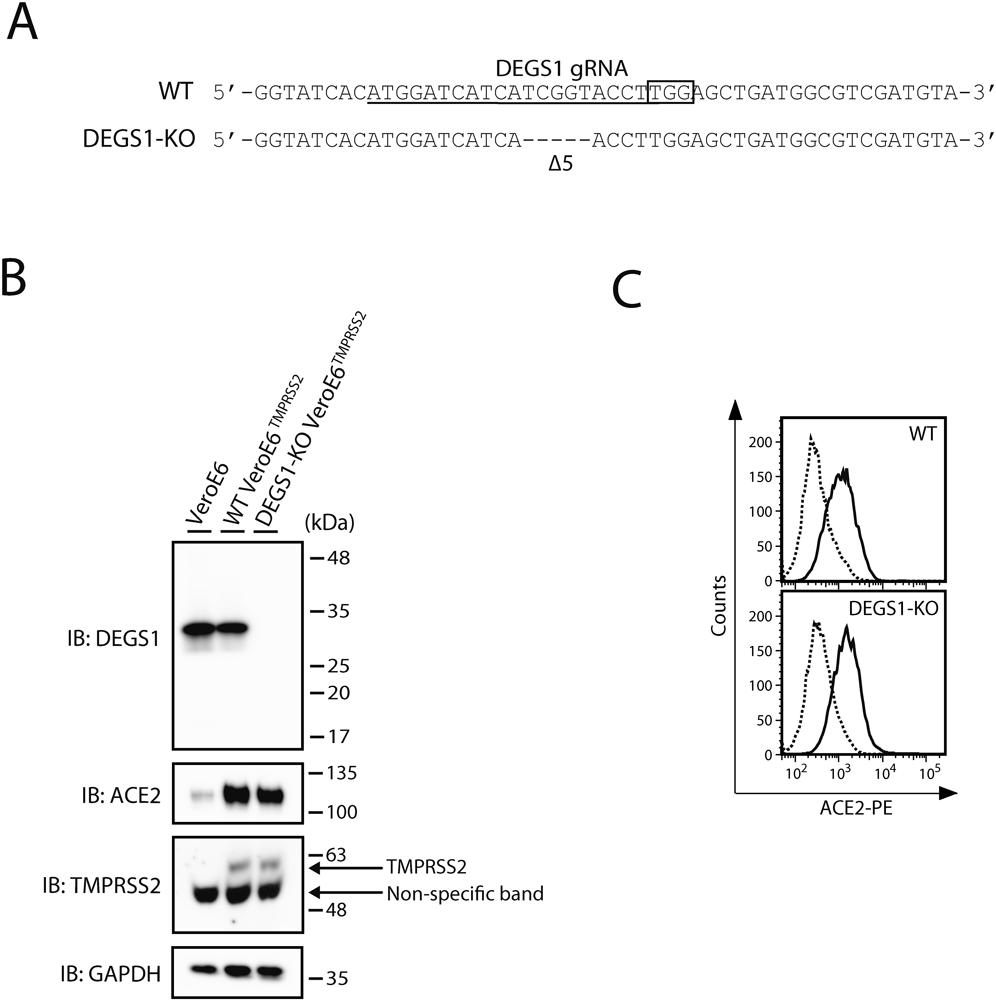
(A) Genomic DNA was extracted from DEGS1-KO cell lines, and a genomic region containing the target sites of the gRNAs was sequenced. The 20-bp target sequence of gRNA is underlined. The sequence of protospacer adjacent motif is boxed. Dashes depict the identified deletions. The number of deletions (Δ, deletions) is shown. (B) Cells were lysed and analyzed by immunoblotting with the indicated antibodies. One representative experiment is shown, and similar results were obtained in three independent experiments. (C) Cell-surface expression level of ACE2 in cells was analyzed by flowcytometry using the anti-ACE2 antibody (thick line) and isotype control (dashed line). Data are from a single representative experiment, and similar results were obtained in three independent experiments.
DEGS1 introduces the 4,5-trans-double bond into DHCer, and produces ceramide (Cer), the core structure of sphingolipids. In mammalian cells, DHCer and Cer are composed of N-acyl chains of various lengths, C16:0, C18:0, C20:0, C22:0, C24:0, and C24:1 (9). To further characterize DEGS1-KO cells, we quantified Cer and DHCer levels in these cells using LC-MS/MS. The levels of all Cer species with a distinct acyl chain, a product of DEGS1, were lower in DEGS1-KO cells compared to WT cells (Figs. 2A–F). In addition, the levels of all DHCer species with a distinct acyl chain, a substrate of DEGS1, were higher in DEGS1-KO cells compared to WT cells (Figs. 2G–L). These results indicate consistent changes in the sphingolipid profile in DEGS1-KO cells.
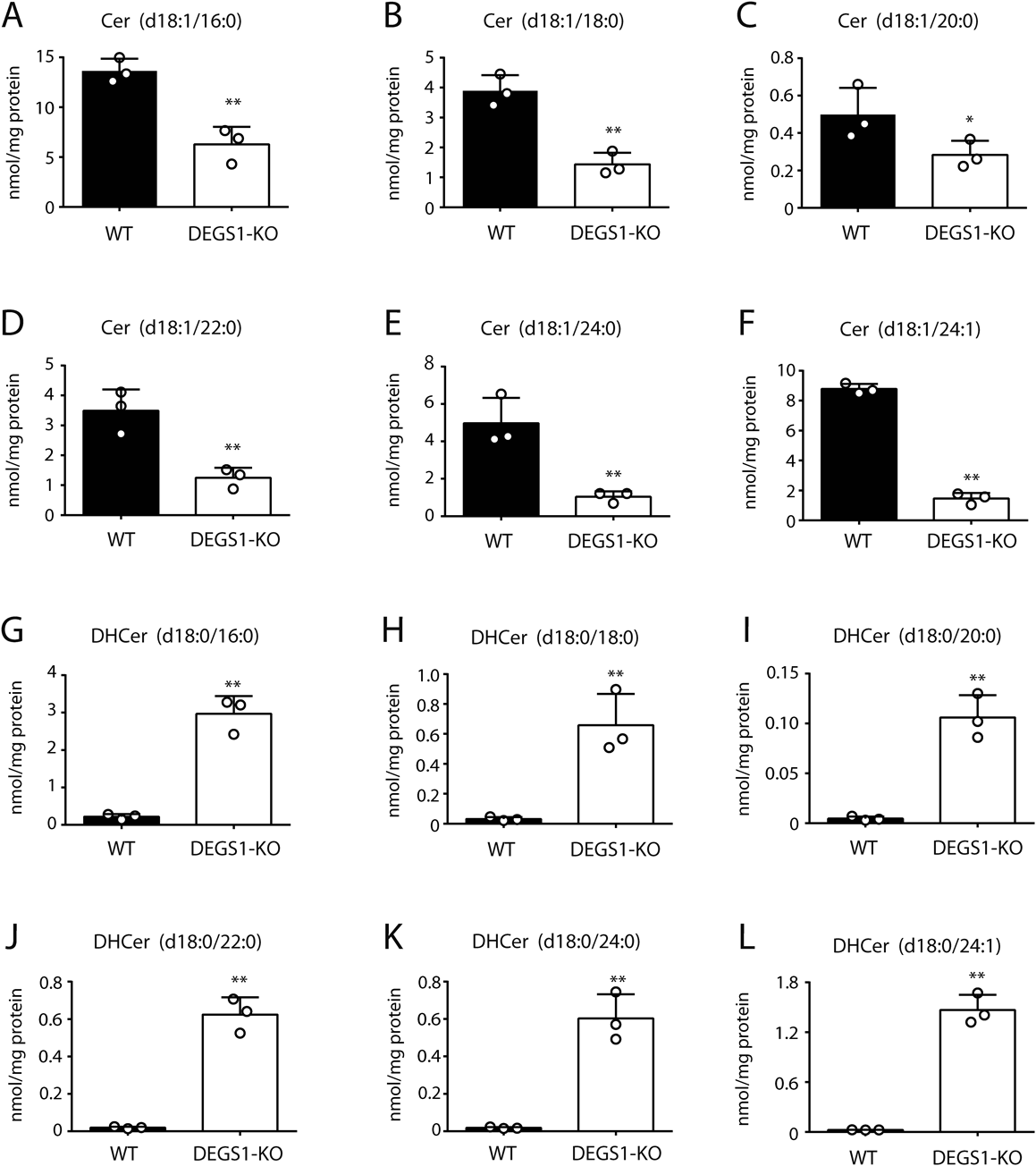
Levels of Cer (A–F) and DHCer (G–L) species with distinct acyl chain in WT and DEGS1-KO cells quantified by LC-MS/MS. Data are mean ± S.D. of three independent experiments. ** p < 0.01, * p < 0.05., by Student t-test.
The effect of DEGS1 disruption on SARS-CoV-2 infection was investigated with RT-qPCR assay (Fig. 3A) and cytopathic effect-based assay (Fig. 3B). First, no significant difference was observed in virus production in the culture supernatant between WT and DEGS1-KO cells (Fig. 3A). Thus, DEGS1 disruption does not seem to affect the life cycles of SARS-CoV-2, such as virus replication and release. Second, the virus-induced cytopathic effect was still observed in DEGS1-KO cells (Fig. 3B). We also examined the antiviral activity of 4-HPR in DEGS1-KO cells to determine the involvement of DEGS1 in SARS-CoV-2 infection. The cytotoxic effect of SARS-CoV-2 infection on DEGS1-KO cells was decreased in a 4-HPR concentration-dependent manner (Fig. 3B). Importantly, the EC50 value of 4-HPR in DEGS1-KO cells (4.1 µM) was almost identical to the value reported previously in WT cells (4.4 µM). These findings indicate that DEGS1 is not involved in SARS-CoV-2 infection, including the replication process and the release of viral progeny.
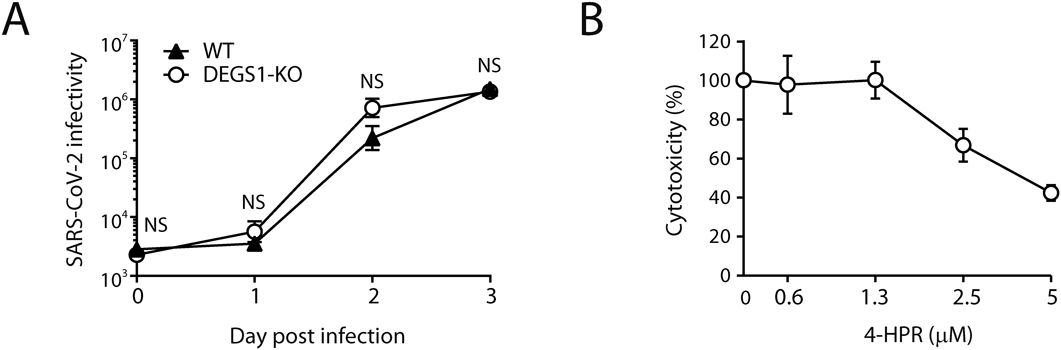
(A) Cells were infected with SARS-CoV-205-2N and supernatants were collected daily. Viral RNA was detected by RT-qPCR. (B) DEGS1-KO cells were treated for 3 d with the indicated concentrations of 4-HPR. Cell viability was examined using the WST-8 assay. Results are normalized to the cell viability in vehicle/DMSO-treated cells. Results are expressed as the percentage of inhibition in compound-treated cells compared to that in the vehicle/DMSO-treated cells. Data are mean ± S.D. of three independent experiments.
In this study, we generated DEGS1-KO VeroE6TMPRSS2 cells with high DHCer levels. We then found that SARS-CoV-2 infected DEGS1-KO cells as efficiently as WT cells. In addition, 4-HPR showed similar anti-viral activity in DEGS1-KO compared with WT cells. These results indicate the lack of involvement of DEGS1 in SARS-CoV-2 infection including the replication process and release of viral progeny. DEGS2, a homologous isoform of DEGS1 as reported to act as a bifunctional enzyme with either C4-monooxygenase activity and Δ4-desaturase activity.10) Cer in DEGS1-KO cells may have been derived from DEGS2.
Sphingolipids are critical in all stages of viral life cycle, such as cell binding in human rhinovirus, entry in influenza virus, replication in hepatitis C virus, and cell lysis and release in adenovirus.11–14) The human immunodeficiency virus (HIV) infection can be suppressed by replacing Cer with DHCer with genetic and pharmacological blockade of DEGS1.15) The same study reported that the increase in dihydrosphingomyelin levels, a DHCer-containing sphingolipid, enforced the rigidity of membrane domains known to be resistant to the insertion of the HIV envelope glycoprotein, with resultant inhibition of HIV infection. What is the reason for the different effect of DHCer level on HIV and SARS-CoV-2 infections? While our study did not examine this issue directly, the localization of viral receptor may explain the difference in the effects of DHCer levels on virus infection. HIV receptor CD4 is predominantly localized in the lipid rafts, sphingolipid-enriched microdomains in the cell membranes,16) whereas SARS-CoV-2 receptor ACE2 is not.6)
4-HPR suppressed SARS-CoV-2 infection in DEGS1-independent mechanism. The antiviral activity of 4-HPR has been confirmed in many viruses, such as HIV, dengue virus, and zika virus.17–19) 4-HPR inhibits the replication of dengue virus by blocking the association of the viral RNA-dependent RNA polymerase (non-structural protein 5) and host’s nuclear transport factors, importin α/β1,18) though the mechanism underlying the inhibition of viral infection by 4-HPR remains unclear. Further investigation is needed to elucidate the exact mechanism underlying the inhibition in SARS-CoV-2 infection by 4-HPR. 4-HPR has been extensively studied for cancer treatment, and exhibited a low-toxicity profile in several clinical trials.20,21) Therefore, the results of our study, along with accumulated clinical data on the safety of 4-HPR, are potential candidates for the treatment of COVID-19.
This work was supported by a Grant from the National Center for Global Health and Medicine (#21A1007).
The authors declare no conflict of interest.