2014 年 62 巻 11 号 p. 1092-1099
2014 年 62 巻 11 号 p. 1092-1099
Eight new hydroperoxides and a new enone of germacrane-type sesquiterpenoids were isolated from the aerial parts of eight different samples of Eupatorium heterophyllum DC. (Asteraceae) collected in P. R. China. The structures were determined based on spectroscopic analyses. Seven of the eight samples produced hiyodorilactone A as a major constituent, while one afforded neither hiyodorilactone nor hydroperoxide. The results indicated the presence of diversity within this species.
Eupatorium s. str. plants (Asteraceae) are distributed in North America, Asia and Europe. In Asia, they occur mainly between an altitude of 1000 and 3000 m.1) Sesquiterpenoids and aromatic compounds have been reported from Eupatorium species.2) Kupchan et al. isolated eupaserrin and deacetyleupaserrin as antileukemic sesquiterpenoids from E. semiserratum in 1973.3) Takahashi et al. reported isolation and biological activities of hiyodorilactones A–F from E. sachalinense.4,5)
We are interested in biologically active terpenoids from Asteraceae plants and have studied the chemical constituents of Ligularia,6–21) Saussurea,22,23) Cremanthodium,24–27) and Eupatorium.28–31) In our previous study on E. glehnii growing in Japan, especially in Obihiro (Hokkaido), Fujimi-cho (Nagano), and Tokushima, a large diversity in the chemical constituents was found, while there was no diversity in DNA sequences (atpB-rbcL intergenic region).30) In our continuous research on the genus Eupatorium, we collected E. heterophyllum DC. (Asteraceae) in P. R. China in 2006 and 2007 (Fig. 1). Five samples, 1–5, were collected in the northwestern part of Yunnan Province, and sample 6 was found in the eastern part of Yunnan Province. The other two samples, 7 and 8, were collected in the southern part of Sichuan Province. Here we report the structures of the chemical constituents isolated from 8 samples of E. heterophyllum and the diversity in these samples.

Solid and dotted lines indicate rivers and boundaries of provinces, respectively. Open circles and filled triangles indicate major cities and peaks, respectively.
EtOAc or MeOH extracts of the aerial part of E. heterophyllum were subjected to silica gel column chromatography and repeated HPLC to yield 22 compounds, 9 of which were new (Fig. 2).
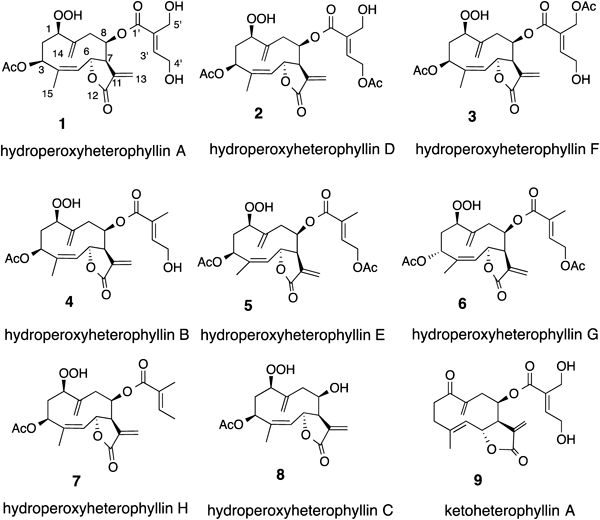
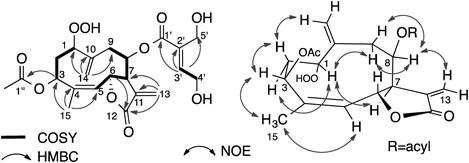
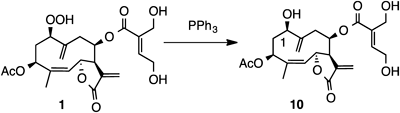
Compound 1 showed a quasimolecular ion peak at m/z 453 and its molecular formula was determined to be C22H28O10 by high resolution (HR)-chemical ionization (CI)-MS and 13C-NMR data. The IR spectrum exhibited absorption at 3500–3200 cm−1 for hydroxy groups and 1740 and 1716 cm−1 for carbonyl groups. The 1H-NMR spectrum indicated the presence of a methyl group attached to the olefinic carbon, an acetyl group, two sets of exomethylene groups, two olefinic protons, four oxymethine protons, and two oxymethylene protons. The 13C-NMR and hetero-nuclear single quantum coherence (HSQC) spectra showed the presence of two methyl, six methylene, seven methine, and seven quaternary carbon atoms. As the degree of unsaturation was nine, this compound should be bicyclic. The correlation spectroscopy (COSY) spectrum clearly showed the proton spin system from H-1 to H-3, and from H-5 to H-9, respectively (Fig. 3). Hetero-nuclear multiple-bond connectivity (HMBC) correlations between H3-15 and C-3, C-4, and C-5, between H2-14 and C-1, C-9, and C-10, between H2-13 and C-7, C-11, and C-12, between H-3 and C-1,” between H-6 and C-12, and between H-3′ and C-1′, C-2′, C-4′, and C-5′ were observed. From these observations, this compound is inferred to be a germacranolide substituted with an acetoxy group at C-3 and a γ-lactone at C-6 and C-12, respectively. The position of the unsaturated ester was suggested by the chemical shift of H-8 (δH 5.21) and C-8 (δC 76.2). The other two oxygen atoms were assigned to a hydroperoxy group at C-1 position, supported by the proton signals at δ 7.97 (1H, br s, OOH) and 4.36 (1H, dd, J=10.5, 3.7 Hz, H-1). The presence of a hydroperoxy group was confirmed by both the starch test and reduction of compound 1 to a 1-hydroxy derivative 10 using PPh3 in benzene (Fig. 4).
The stereochemistry was determined by the nuclear Overhauser effect correlated spectroscopy (NOESY) spectrum. Because NOE between H-5 and H3-15 was observed, the double bond at C-4 and C-5 should be Z. Although the coupling constant between H-6 and H-7 was 1.7 Hz, these protons were trans to each other in the γ-butylolactone ring. These phenomena were observed in other germacranolides.29,30) The unsaturated ester was 4′,5′-dihydroxytiglate, because of the presence of NOE between H-4′ and H-5′. The NOE between H-1 and H-7, as well as other NOEs shown in Fig. 3, supported the stereochemistry depicted in the formula. Therefore, the whole structure was established as depicted in the figure and was named hydroperoxyheterophyllin A.
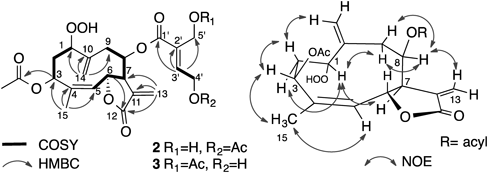
Spectroscopic data of compounds 2 and 3 were very similar. Both IR spectra exhibited the absorption of hydroxy and carbonyl groups and HR-FAB-MS indicated the same molecular formula, C24H30O11. The 1H-NMR spectrum showed the presence of three methyl groups, two of which were acetyl groups, two sets of exomethylene groups, two olefinic protons, four oxymethine protons, and two oxymethylene protons. The 13C-NMR and HSQC spectra showed the presence of three methyl, six methylene, seven methine, and eight quaternary carbon atoms. In addition to the same HMBC correlations as observed in compound 1, the correlation to acetyl carbonyl carbon was observed from H2-4′ (δ 4.81 and 4.85) in compound 2, whereas from H2-5′ (δ 4.79 and 4.92) in compound 3 (Fig. 5), showing that compounds 2 and 3 were O-acetyl derivatives of compound 1 at C-4′ and C-5′, respectively. The NOE correlations were almost the same as those of compound 1. Therefore, the structures of compounds 2 and 3 were established as depicted in the formula and were named hydroperoxyheterophyllins D and F, respectively.
Compound 4, showing a positive starch test, exhibited a quasimolecular ion peak at m/z 437 and the molecular formula was determined to be C22H28O9 by HR-FAB-MS and 13C-NMR data. The 1H- and 13C-NMR spectra are very similar to those of compound 1, except that there is an additional methyl signal at δ 1.80 and the absence of oxymethylene proton signals at δ 4.34 in compound 1. These observations and the two dimensional (2D)-NMR analyses suggested that compound 4 should be a 5′-deoxy derivative of compound 1 (Fig. 6). The stereochemistry was determined from NOESY data. The geometry of the ester part was determined to be E, because the NOE between H2-4′ and H3-5′ was observed. Compound 4 was named hydroperoxyheterophyllin B.
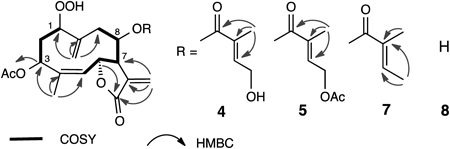
The molecular formula of compound 5 was determined to be C24H30O10 by HR-FAB-MS and 13C-NMR data. Both 1H- and 13C-NMR data were very similar to those of compound 4 (Tables 1, 2). However, there was an additional acetyl proton at δ 2.07 in compound 5, which was supported by the MS and 13C-NMR data (Table 2). The position of this acetoxy group was suggested to be at C-4′, because the proton signal at δ 4.35 (H-4′) in compound 4 shifted to δ 4.72 in compound 5. The geometry of the double bond was retained as 2′E, because the NOE between H2-4′ and H3-5′ was observed. Compound 5 was named hydroperoxyheterophyllin E.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4.36 (1H, dd, 10.5, 3.7) | 4.36 (1H, dd, 9.7, 4.4) | 4.37 (1H, dd, 9.8, 4.7) | 4.37 (1H, dd, 11.2, 4.1) | 4.37 (1H, dd, 10.7, 3.5) | 4.04 (1H, dd, 12.0, 3.4) | 4.37 (1H, dd, 10.6, 3.8) | 4.33 (1H, dd, 10.8, 3.3) | — | 4.06 (1H, ddd, 11.5, 8.6, 3.0) |
| 2 | 2.52 (1H, m) | 2.49 (2H, m) | 2.49 (2H, m) | 2.48 (2H, m) | 2.49 (2H, m) | 2.53 (1H, ddd, 12.4, 12.0, 4.2) | 2.49 (2H, m) | 2.48 (1H, td, 10.8, 2.7) | 3.31 (1H, br s) | 2.43 (1H, br dd, 15.7, 11.5) |
| 2.46 (1H, m) | — | — | — | — | 2.28 (1H, ddd, 12.4, 12.0, 3.4) | — | 2.43 (1H, ddd, 10.8, 5.1, 3.3) | 2.38 (1H, br s) | 2.21 (1H, ddd, 15.7, 5.7, 3.0) | |
| 3 | 5.41 (1H, dd, 4.9, 3.2) | 5.41 (1H, dd, 4.6, 2.9) | 5.41 (1H, dd, 4.5, 3.3) | 5.40 (1H, dd, 5.1, 2.7) | 5.40 (1H, dd, 5.1, 2.7) | 5.75 (1H, dd, 12.0, 4.2) | 5.40 (1H, dd, 4.9, 2.7) | 5.39 (1H, dd, 5.1, 2.7) | 2.60 (1H, br s) | 5.35 (1H, dd, 5.7, 2.4) |
| — | — | — | — | — | — | — | — | 2.37 (1H, br s) | — | |
| 5 | 5.34 (1H, dq, 10.5, 1.5) | 5.34 (1H, dq, 10.5, 1.5) | 5.33 (1H, dq, 10.6, 1.4) | 5.34 (1H, dq, 10.5, 1.5) | 5.34 (1H, dq, 10.5, 1.5) | 5.34 (1H, d, 11.0) | 5.34 (1H, dq, 10.5, 1.6) | 5.33 (1H, dq, 10.4, 1.5) | 5.11 (1H, br d, 9.2) | 5.33 (1H, dq, 9.6, 1.4) |
| 6 | 6.15 (1H, dd, 10.5, 1.7) | 6.13 (1H, dd, 10.5, 1.7) | 6.13 (1H, dd, 10.6, 1.6) | 6.16 (1H, dd, 10.5, 2.1) | 6.13 (1H, dd, 10.5, 2.0) | 5.64 (1H, dd, 11.0, 2.5) | 6.13 (1H, dd, 10.5, 2.2) | 5.95 (1H, dd, 10.4, 2.3) | 4.94 (1H, br t, 9.2) | 6.08 (1H, br d, 9.6) |
| 7 | 3.13 (1H, m) | 3.12 (1H, m) | 3.11 (1H, m) | 3.09 (1H, m) | 3.09 (1H, br s) | 3.01 (1H, br s) | 3.07 (1H, m) | 2.95 (1H, m) | 2.89 (1H, br s) | 3.15 (1H, br s) |
| 8 | 5.21 (1H, m) | 5.22 (1H, m) | 5.26 (1H, m) | 5.18 (1H, m) | 5.19 (1H, m) | 5.19 (1H, m) | 5.17 (1H, m) | 4.11 (1H, m) | 5.73 (1H, br d, 6.5) | 5.23 (1H, m) |
| 9 | 2.96 (1H, dd, 15.0, 5.1) | 2.97 (1H, dd, 15.2, 4.9) | 2.95 (1H, dd, 15.1, 4.9) | 2.95 (1H, dd, 15.1, 4.7) | 2.95 (1H, dd, 14.9, 5.2) | 2.97 (1H, dd, 15.1, 4.4) | 2.96 (1H, dd, 15.3, 5.5) | 2.73 (1H, dd, 14.7, 5.5) | 3.15 (1H, br d, 14.9) | 2.93 (1H, dd, 14.9, 5.7) |
| 2.57 (1H, dd, 15.0, 2.8) | 2.58 (1H, dd, 15.2, 2.6) | 2.57 (1H, dd, 15.1, 3.0) | 2.54 (1H, dd, 15.1, 2.9) | 2.54 (1H, dd, 14.9, 2.7) | 2.47 (1H, dd, 15.1, 2.9) | 2.49 (1H, m) | 2.54 (1H, dd, 14.7, 3.0) | 2.66 (1H, br dd, 14.9, 6.5) | 2.65 (1H, dd, 14.9, 3.1) | |
| 13 | 6.38 (1H, d, 2.2) | 6.38 (1H, d, 2.0) | 6.37 (1H, d, 2.1) | 6.36 (1H, d, 2.2) | 6.37 (1H, d, 1.9) | 6.39 (1H, d, 2.2) | 6.36 (1H, d, 1.9) | 6.41 (1H, d, 2.1) | 6.27 (1H, d, 3.3) | 6.37 (1H, d, 1.7) |
| 5.83 (1H, d, 1.8) | 5.82 (1H, d, 1.7) | 5.82 (1H, d, 2.0) | 5.80 (1H, d, 2.0) | 5.80 (1H, d, 1.9) | 5.79 (1H, d, 2.0) | 5.79 (1H, d, 1.9) | 5.74 (1H, d, 2.1) | 5.60 (1H, s) | 5.82 (1H, d, 2.0) | |
| 14 | 5.52 (1H, s) | 5.51 (1H, s) | 5.51 (1H, s) | 5.51 (1H, s) | 5.50 (1H, s) | 5.81 (1H, s) | 5.49 (1H, s) | 5.55 (1H, s) | 5.96 (1H, s) | 5.43 (1H, s) |
| 5.37 (1H, s) | 5.37 (1H, s) | 5.35 (1H, s) | 5.34 (1H, s) | 5.32 (1H, s) | 5.41 (1H, s) | 5.31 (1H, s) | 5.50 (1H, s) | 5.60 (1H, s) | 5.19 (1H, s) | |
| 15 | 1.87 (3H, d, 1.5) | 1.87 (3H, d, 1.5) | 1.87 (3H, d, 1.4) | 1.87 (3H, d, 1.5) | 1.87 (3H, d, 1.5) | 1.86 (3H, s) | 1.87 (3H, d, 1.6) | 1.84 (3H, d, 1.4) | 1.85 (3H, d, 0.7) | 1.85 (3H, d, 1.4) |
| 3′ | 6.95 (1H, t, 5.7) | 6.80 (1H, t, 6.3) | 7.06 (1H, t, 5.9) | 6.83 (1H, tq, 5.8, 1.4) | 6.75 (1H, t, 6.1) | 6.75 (1H, t, 6.1) | 6.87 (1H, qq, 7.1, 1.3) | — | 6.92 (1H, t, 5.7) | 6.96 (1H, t, 5.7) |
| 4′ | 4.42 (2H, d, 5.7) | 4.85 (1H, dd, 14.7, 6.3) | 4.46 (2H, d, 5.9) | 4.35 (2H, m) | 4.72 (2H, d, 6.1) | 4.74 (2H, d, 6.1) | 1.77 (3H, dq, 7.1, 1.3) | — | 4.49 (1H, dd, 15.4, 5.7) | 4.43 (1H, dd, 5.7, 4.1) |
| — | 4.81 (1H, dd, 14.7, 6.3) | — | — | — | — | — | — | 4.45 (1H, dd, 15.4, 5.7) | — | |
| 5′ | 4.34 (2H, d, 5.3) | 4.37 (1H, d, 12.9) | 4.92 (1H, d, 12.1) | 1.80 (3H, d, 1.4) | 1.85 (3H, s) | 1.86 (3H, s) | 1.80 (3H, quint, 1.3) | — | 4.38 (1H, s) | 4.36 (1H, dd, 12.7, 5.5) |
| — | 4.31 (1H, d, 12.9) | 4.79 (1H, d, 12.1) | — | — | — | — | — | — | 4.32 (1H, dd, 12.7, 5.9) | |
| 3-OAc | 2.09 (3H, s) | 2.09 (3H, s) | 2.08 (3H, s) | 2.08 (1H, s) | 2.069 (3H, s) | 2.11 (3H, s) | 2.06 (3H, s) | 2.07 (3H, s) | — | 2.09 (1H, s) |
| 4′-OAc | — | 2.08 (3H, s) | — | — | 2.074 (3H, s) | 2.12 (3H, s) | — | — | — | — |
| 5′-OAc | — | — | 2.07 (3H, s) | — | — | — | — | |||
| OOH | 7.97(1H, br s) | 8.14 (1H, s) | 7.96 (1H, s) | 7.83 (1H, s) | 7.83 (1H, s) | 7.70 (1H, s) | — | 7.79 (1H, s) | — | — |
| 1-OH | — | — | — | — | — | — | — | — | — | 1.33 (1H, d, 8.6) |
| 4′-OH | — | — | — | — | — | — | — | — | — | 2.26 (1H, br s) |
| 5′-OH | — | — | — | — | — | — | — | — | — | 2.48 (1H, br s) |
| 8-OH | — | — | — | — | — | — | — | 1.81 (1H, br d, 3.7) | — | — |
| 1 | 2 | 3 | 4 | 5 | 6 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|
| 1 | 84.0 | 84.1 | 84.2 | 84.2 | 84.2 | 83.7 | 84.4 | 203.6 |
| 2 | 29.9 | 30.0 | 30.1 | 30.0 | 30.0 | 31.4 | 30.2 | 34.3 |
| 3 | 73.0 | 73.0 | 73.0 | 73.0 | 73.1 | 69.2 | 72.8 | 38.1 |
| 4 | 138.9 | 138.5 | 138.4 | 138.3 | 138.2 | 136.9 | 137.4 | 144.2 |
| 5 | 125.7 | 126.2 | 126.3 | 126.3 | 126.4 | 126.6 | 126.8 | 125.1 |
| 6 | 73.9 | 73.3 | 73.3 | 73.4 | 73.2 | 72.1 | 72.8 | 74.7 |
| 7 | 47.4 | 47.5 | 47.6 | 47.7 | 47.6 | 48.4 | 49.1 | 51.4 |
| 8 | 76.2 | 76.4 | 76.0 | 75.7 | 75.9 | 76.3 | 73.7 | 68.5 |
| 9 | 37.5 | 37.8 | 37.9 | 37.9 | 37.9 | 38.2 | 41.3 | 30.3 |
| 10 | 137.1 | 136.7 | 136.9 | 137.0 | 136.9 | 136.1 | 137.0 | 141.1 |
| 11 | 136.8 | 136.8 | 136.8 | 136.8 | 136.8 | 136.9 | N/D | 135.9 |
| 12 | 170.3 | 169.4 | 169.5 | 169.7 | 169.4 | 168.9 | 169.8 | 169.3 |
| 13 | 125.4 | 125.3 | 125.2 | 125.0 | 125.1 | 125.1 | 123.7 | 121.2 |
| 14 | 122.1 | 122.5 | 122.4 | 122.4 | 122.4 | 122.7 | 122.0 | 127.9 |
| 15 | 22.8 | 23.0 | 23.0 | 23.0 | 23.0 | 17.7 | 23.0 | 17.2 |
| 1′ | 165.6 | 165.2 | 164.7 | 166.1 | 165.7 | 166.0 | — | 165.6 |
| 2′ | 130.9 | 133.1 | 126.6 | 127.4 | 129.6 | 129.7 | — | 131.6 |
| 3′ | 145.5 | 139.0 | 148.0 | 142.1 | 136.5 | 136.9 | — | 144.4 |
| 4′ | 58.7 | 60.4 | 59.6 | 59.8 | 61.0 | 60.9 | — | 59.2 |
| 5′ | 56.5 | 57.0 | 57.9 | 12.5 | 12.7 | 12.8 | — | 57.2 |
| 3-OAc | 170.0 | 170.7 | 171.4 | 169.7 | 169.5 | 170.1 | 169.8 | — |
| 20.9 | 20.7 | 20.9 | 21.0 | 21.0 | 20.8 | 21.1 | — | |
| 4′-OAc | — | 169.6 | — | — | 170.6 | 170.8 | — | — |
| — | 20.9 | — | — | 20.7 | 21.1 | — | — | |
| 5′-OAc | — | — | 169.7 | — | — | — | — | — |
| — | — | 21.0 | — | — | — | — | — |
The molecular formula of compound 7 was determined to be C22H28O8 and the spectroscopic features were very similar to those of compound 4, except that there was one more methyl signal at δ 1.77 and the absence of a hydroxymethylene group in compound 4. Compound 7 was also positive in the starch test. The COSY spectrum suggested that the unsaturated ester part was either an angeloyl or a tigloyl group. However, the chemical shifts of H-3′ (δ 6.87) indicated that this is a tiglate group, which is supported by the NOESY spectrum. The other part was the same as those of compound 4. Therefore, compound 7 was determined to be a deoxygenated derivative of compound 4 as depicted in the formula and it was named hydroperoxyheterophyllin H.
Compound 8 showed a quasimolecular ion peak at m/z 361 [M+Na]+ (FAB) and the molecular formula was determined to be C17H22O7 by HR-FAB-MS and 13C-NMR data. The 1H-NMR spectrum showed the presence of a methyl group attached to the sp2 carbon, an acetyl group (δ 2.07), an exomethylene group, an olefinic proton, and four oxymethine protons. The presence of a γ-butylolactone was suggested by the IR absorption at 1737 cm−1. However, signals due to an ester substituted at C-8 in the compounds discussed above were not detected. The oxymethine proton at C-8 was detected at δ 4.11, shifted to a higher field, suggesting that a hydroxy group was substituted. This assumption was supported by 2D-NMR analysis. This new compound was named hydroperoxyheterophyllin C.
Compound 6 had the same molecular formula, C24H30O10, as that of 5. The 1H- and 13C-NMR data of compound 6 were similar to those of compound 5, but H-1, H2-2, and H-3 were different (Tables 1, 2). The difference in the chemical shift at H-6 was large; δ 6.13 in compound 5, while δ 5.64 in compound 6. This phenomenon implied that the substituent was in proximity to H-6; namely, the acetoxy group at C-3 had presumably different stereochemistry. This assumption was revealed by the observation of NOE between H-6 and H-3 as well as between H3-15 and H-5 (Fig. 7). The 13C-NMR signal for C-15 of compound 6 resonated at δC 17.7, which was in the higher field than those of compounds 1–5 (Table 2). This phenomenon was also explained by proximity of the acetoxy group at C-3 to the methyl group at C-4. Therefore, this compound was established as depicted in the formula and was named hydroperoxyheterophyllin G.
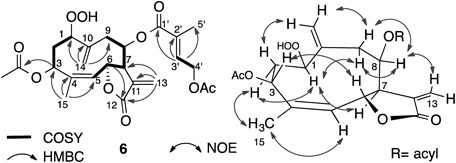
Compound 9 showed a molecular ion peak at m/z 376 and its molecular formula was determined to be C20H24O7 by HR-electron ionization (EI)-MS and 13C-NMR data. The IR spectrum exhibited absorption at 3500–3300, 1740, 1712, and 1650 cm−1, indicating the presence of a hydroxy, a lactone, and an α,β-unsaturated carbonyl group. The 13C-NMR spectrum showed the presence of a methyl group attached to the sp2 carbon, seven methylenes, two of which were oxymethylene, five methines, two of which were oxymethine, and seven quaternary carbons, three of which were carbonyl groups. Therefore, the degree of unsaturation was nine and this molecule should be bicyclic. The COSY correlations from H2-2 to H2-3, and from H-6 to H2-9 were revealed. Long-range correlations between H3-15 and C-3, C-4, and C-5, between H2-13 and C-11, C-12, and C-7, between H2-14 and C-1, C-9, and C-10, between H-8 and C-6, C-10, and C-1′ were observed. Therefore, this compound had a germacrane skeleton with the γ-butylolactone at C-6 and C-12, and C-1 was a carbonyl carbon. Because the NOESY spectrum showed correlations between H-5 and H-3α and H-7α and between H-6 and H3-15, the conformation and configuration of a ten-membered ring should be as shown in Fig. 8. Compound 9 was named ketoheterophyllin A.
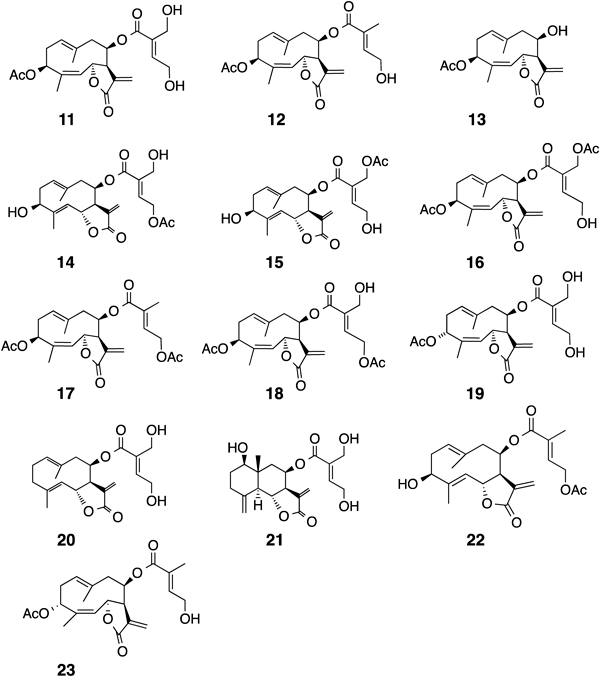
Other compounds, 11–23, were identified by comparing their spectroscopic and physicochemical data with those reported and independent analyses (Fig. 9). Samples 1, 4, and 7 contained quite a lot of hydroperoxyheterophyllin A (1) (1.2, 1.4, and 1.6% of the extract). The most abundant compound was hiyodorilactone A (11),4) except in sample 6. Sample 6 produced four compounds, 9, 19, 20, and 21, which were not isolated from other samples (Table 3). 3-Deoxygenated compounds were characteristic of sample 6, suggesting that the sample belonged to a different chemotype.32) Sample 6 was geographically isolated from the other samples.

| Sample | Compound (mg)a) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | |
| 1 | 12 | 213 | 31 | 27 | 2 | 5 | 1 | 2 | ||||||||||||||
| 2 | 1 | 1 | 2 | 313 | 140 | 25 | 4 | 2 | 4 | |||||||||||||
| 3 | 192 | 33 | 18 | 3 | 10 | |||||||||||||||||
| 4 | 13 | 1 | 5 | 127 | 24 | 1 | 5 | |||||||||||||||
| 5 | 2 | 1 | 1 | 1 | 160 | 64 | 19 | 3 | ||||||||||||||
| 6 | 6 | 4 | 7 | 4 | ||||||||||||||||||
| 7 | 21 | 4 | 2 | 2 | 56 | 13 | 9 | |||||||||||||||
| 8 | 86 | 23 | ||||||||||||||||||||
a) Weights less than 1 mg were shown as 1.
Previous examples of identifying hydroperoxides of a germacrane type33–38) as well as antimicrobial and nuclear factor-kappa B (NF-κB) inhibitory activity38–40) have been reported. Therefore, cytotoxicity was tested for hydroperoxyheterophyllin A (1), hiyodorilactone A (11), hiyodorilactone B (12), hiyodorilactone D (14), and eupatoriopicrin (20) on MiaPaCa-2 and AsPC-1 cell lines. Compounds 11, 12, and 20 showed weak effects on MiaPaCa-2 at 10 µM with 60–70% decreases, respectively, and their IC50 were 7.3, 6.95, and 4.41 µM. Compound 20 showed weak activity on AsPC-1 at 10 µM with 60% decrease (IC50 6.28 µM). Then, inhibition of infection and growth of tripanosoma for compounds 1 and 8 was tested; however, no effect was observed at 10 µM.
ConclusionWe have isolated eight new hydroperoxides of a germacrane type with various acyl groups substituted at C-8 and a new germacranolide enone as well as 13 known related compounds from the aerial parts of eight samples of E. heterophyllum collected in Yunnan and Sichuan provinces, China. Aromatic compounds were previously isolated from the roots of this plant, while the aerial part affords sesquiterpenoids, mainly hiyodorilactone A (11). From this point, E. heterophyllum in China is somehow similar to E. glehnii growing in Hokkaido and Tokushima in Japan. One sample afforded different products from the other samples; thus, intra-specific diversity of chemical constituents is likely to occur in this species, which will be further investigated in the future.
ExperimentalGeneral Procedures: GeneralSpecific rotations and circular dichroism (CD) spectra were measured on a JASCO DIP-1030 and a JASCO J-725 auto recording polarimeter; IR spectra, on a SHIMADZU FT/IR-8400S spectrophotometer (samples were absorbed on a powdered KBr surface and measured by the diffusion reflection method); 1H- and 13C-NMR spectra, on a Varian 400-MR (400 MHz and 100 MHz, respectively) spectrometer; Mass spectra were recorded on a JEOL JMS-700 MStation. Chemcopak Nucleosil 50–5 (4.6×250 mm), COSMOSIL 5SL-II (10×250 mm), and/or TSK-gel G1000HHR (7.8×300 mm) were used for HPLC (JASCO pump system) with a solvent system of n-hexane–EtOAc in different ratios. Silica gel 60 (70–230 mesh; Fuji Silysia and silica gel 60 F254 plates (Merck) were used for the column and TLC, respectively.
Plant MaterialThe samples of E. heterophyllum DC. (Asteraceae) were collected in August 2006 and 2007 at the locations shown in Fig. 1. Voucher specimens were deposited in the Herbarium of Kunming Institute of Botany, Kunming, China (specimen Nos.: 2006–24, 2006–29, 2006–34, 2006–60, 2006–64, 2006–94, 2007–34, and 2007–77). The plants were identified by Dr. Takayuki Kawahara, one of the authors.
Extraction and IsolationThe aerial parts of E. heterophyllum were dried, ground, and then extracted with MeOH or EtOAc to afford extracts. The compounds therein were separated by silica gel column chromatography (n-hexane/EtOAc) followed by HPLC (n-hexane/EtOAc) to afford each compound.
Sample 1 (specimen No.; 2006-24) was extracted with EtOAc to afford an extract (961.7 mg). This extract was separated by silica gel column chromatography followed by HPLC to give 1 (12.2 mg), 11 (213.3 mg),4) 12 (30.9 mg),4) 14 (27.2 mg),5) 15 (1.8 mg),5) 18 (4.6 mg),41) 22 (1.3 mg),42) 23 (1.8 mg).43)
Sample 2 (specimen No.; 2006-29) was extracted with MeOH to afford an extract (938.6 mg). This extract was separated by silica gel column chromatography followed by HPLC to give 5 (1.1 mg), 6 (0.3 mg), 7 (1.6 mg), 11 (313 mg), 12 (140.2 mg), 14 (24.5 mg), 15 (3.9 mg),5) 16 (1.9 mg),5) and 17 (4.3 mg).44)
Sample 3 (specimen No.; 2006-34) was extracted with MeOH to afford an extract (943.2 mg). This extract was separated by silica gel column chromatography followed by HPLC to give 11 (191.9 mg), 12 (33.1 mg), 14 (17.9 mg), 16 (2.7 mg), and 18 (10.0 mg).
Sample 4 (specimen No.; 2006-60) was extracted with MeOH to afford an extract (962.4 mg). This extract was separated by silica gel column chromatography followed by HPLC to give 1 (12.6 mg), 4 (1.3 mg), 8 (4.7 mg), 11 (126.5 mg), 12 (23.6 mg), 13 (1.4 mg),4) and 14 (5.1 mg).
Sample 5 (specimen No.; 2006-64) was extracted with MeOH to afford an extract (894.6 mg). This extract was separated by silica gel column chromatography followed by HPLC to give 1 (2.0 mg), 5 (0.7 mg), 6 (0.2 mg), 7 (0.5 mg), 11 (160.2 mg), 12 (64.0 mg), 14 (18.6 mg), and 17 (2.7 mg).
Sample 6 (specimen No.; 2006-94) was extracted with MeOH to afford an extract (2930.6 mg). This extract was separated by silica gel column chromatography followed by HPLC to give 9 (5.6 mg), 19 (4.3 mg),45) 20 (6.6 mg),46) and 21 (4.2 mg).47)
Sample 7 (specimen No.; 2007-34) was extracted with EtOAc to afford an extract (1279.4 mg). This extract was separated by silica gel column chromatography followed by HPLC to give 1 (20.8 mg), 2 (3.7 mg), 3 (2.2 mg), 4 (2.3 mg), 11 (55.5 mg), 16 (12.8 mg), and 18 (9.0 mg).
Sample 8 (specimen No.; 2007-77) was extracted with MeOH to afford an extract (1036.4 mg). This extract was separated by silica gel column chromatography followed by HPLC to give 11 (86.0 mg) and 12 (23.3 mg).
Compound 1: FT-IR cm−1: 3500−3200, 1740, 1716. FAB-MS m/z: 453.1774 [M+H]+ (Calcd for C22H29O10: 453.1761). MS m/z: 453 [M+H]+. [α]D21 −19.9 (c=0.7, CHCl3). CD (EtOH) [θ] (nm): +23129 (239), −43712 (222), +56958 (204).
Compound 2: FT-IR cm−1: 3500−3200, 1742, 1715. FAB-MS m/z: 517.1663 [M+Na]+ (Calcd for C24H30O11Na: 517.1686). MS m/z: 517 [M+Na]+, 89 (base). [α]D23 −6.9 (c=0.4, EtOH). CD (EtOH) [θ] (nm): +5700 (237), −36000 (217).
Compound 3: FT-IR cm−1: 3500−3200, 1745, 1730, 1713. FAB-MS m/z: 495.1877 [M+H]+ (Calcd for C24H31O11: 495.1866). MS m/z: 495 [M+H]+, 176 (base). [α]D23 −17.2 (c=0.3, EtOH). CD (EtOH) [θ] (nm): +4200 (242), −52000 (211).
Compound 4: FT-IR cm−1: 3500−3200, 1740. FAB-MS m/z: 437.1840 [M+H]+ (Calcd for C22H29O9: 437.1811). MS m/z: 437 [M+H]+, 185, 149, 93 (base), 75. [α]D22 −13.8 (c=0.1, CHCl3). CD (EtOH) [θ] (nm): +10046 (239), −49295 (208), +9620 (201).
Compound 5: FT-IR cm−1: 3500−3200, 1745, 1730, 1715. FAB-MS m/z: 479.1927 [M+H]+ (Calcd for C24H31O10: 479.1917). MS m/z: 479 [M+H]+, 89 (base). [α]D25 −62.0 (c=0.3, EtOH). CD (EtOH) [θ] (nm): +3050 (255), −74000 (212).
Compound 6: FT-IR cm−1: 3500−3200, 1745, 1730, 1715. FAB-MS m/z: 479.1926 [M+H]+ (Calcd for C24H31O10: 479.1917). MS m/z: 479 [M+H]+, 77 (base). [α]D24 +5.6 (c=0.1, EtOH). CD (EtOH) [θ] (nm): +4500 (236), −24000 (213).
Compound 7: FT-IR cm−1: 3400−3200, 1740, 1712, 1650. FAB-MS m/z: 421.1878 [M+H]+ (Calcd for C22H29O8: 421.1862). MS m/z: 421 [M+H]+, 89 (base). [α]D23 −33.2 (c=0.1, CHCl3). CD (EtOH) [θ] (nm): +11683 (240), −70708 (207), −8582 (200).
Compound 8: FT-IR cm−1: 3500−3300, 1737. FAB-MS m/z: 361.1256 [M+Na]+ (Calcd for C17H22O7Na: 361.1263). MS m/z: 361 [M+Na]+, 89 (base). [α]D22 −14.5 (c=0.1, CHCl3). CD (EtOH) [θ] +6586 (265), −22954 (217), +40234 (201).
Compound 9: FT-IR 3500−3300, 1740, 1712, 1650 cm−1. EI-MS Obs m/z 376.1535 (M+) (Calcd for C20H24O7 376.1522). MS m/z 376 (M+), 262, 244, 97 (base), 69, 41. [α]D21 +109.9 (c=0.5, CHCl3). CD (EtOH) [θ] (nm): +4533 (295), +28085 (240), −5579 (208).
Reduction of Hydroperoxy Group to Hydroxy GroupA solution of compound 1 (1.5 mg) in PhH (1 mL) was treated with PPh3 (1.3 mg) at rt with stirring overnight. The mixture was purified by silica gel column chromatography (CHCl3–AcOEt) to give compound 10 (0.2 mg). EI-MS m/z 436 (M+), 305, 244, 97 (base).
We thank Prof. Shinzaburo Takamiya, Faculty of Medical Sciences, Juntendo University, for evaluation of the inhibitory activity of tripanosoma, and Prof. Hachiro Sugimoto, FarmaEight, Kyoto, for cytotoxicity.