2016 年 64 巻 1 号 p. 59-62
2016 年 64 巻 1 号 p. 59-62
Cristazine (1), a new class of dioxopiperazine alkaloid, along with previously isolated chetomin (2), neoechinulin A (3), and golmaenone (4), were isolated from the mudflat-sediment-derived fungus Chaetomium cristatum. The structure and absolute stereochemistry of 1 was assigned on the basis of NMR, electron impact (EI)-MS, tandem FAB-MS/MS, and circular dichroism (CD) experiments. Compounds 1–4 displayed potent radical-scavenging activity against 2,2-diphenyl-1-picrylhydrazyl (DPPH), with IC50 values of 19, 15, 24, and 20 µM, respectively, which were similar to that of the positive control, ascorbic acid (IC50, 20 µM). Compound 1 also displayed cytotoxic activity against human cervical carcinoma (HeLa) cells, with an IC50 value of 0.5 µM.
Marine microorganisms, particularly marine fungi, have recently gained prominence as an important source of biologically active secondary metabolites.1,2) Among the marine fungi, those associated with marine sediments are a particularly promising source of novel natural products because they inhabit a special ecological niche.2,3) As part of a program to explore the bioactive metabolites produced by fungi isolated from marine habitats, we directed our efforts towards the microorganisms associated with the sediments of marine mudflats,4) and isolated the fungal strain Chaetomium cristatum. This paper describes the isolation and structural characterization of a new dioxopiperazine alkaloid, cristazine (1), and the known chetomin (2),5,6) neoechinulin A (3),7,8) and golmaenone (4)7) from C. cristatum.
Cristazine (1), [α]D20 +220 (c=0.3, CHCl3), was isolated as a colorless amorphous solid, and lacked the molecular ion in the electron impact (EI), FAB or electrospray ionization (ESI) mass spectra, displaying peaks at m/z 43 [COCH3]+ in the EI-MS and m/z 646 [M+H−COCH3]+ in the FAB-MS (see Figs. S1 and S4 in the supplementary information). The mass spectral fragmentation patterns demonstrated fission of the C-3/12 bond, the side chain being of the formula COCH3 (see Fig. S3 in the supplementary information). The compound had isotopic clusters at m/z 646 [M (35Cl)]+ (11.8) and 648 [M (37Cl)]+ (3.6) with a ratio of about 3 : 1 in the EI-MS, suggesting the presence of a single chlorine atom. The IR spectrum of 1 showed bands characteristic of amine (3484, 3355 cm−1), amide (1681 cm−1), and aromatic (1608 cm−1) functionalities. The presence of a broad amide band in the IR spectrum and the presence of a positive Cotton effect at 238 nm (Δε +31.9) indicated the presence of a dioxopiperazine moiety9,10) in compound 1. The Cotton effects at 269 nm and 303 nm were assigned to a sulfide chromophore,10) the presence of which was supported by the fragment ion m/z 253 [m/z 285−S]+ in the fragmenting pattern of compound 1 (see Figs. S1 and S3 in the supplementary information). Detailed analysis of the 1H- and 13C-NMR spectra of 1, including the results of distortionless enhancement by polarization transfer (DEPT), correlation spectroscopy (COSY), heteronuclear multiple quantum correlation (HMQC), and heteronuclear multiple bond coherence (HMBC) experiments, revealed diagnostic signals for two different indolyl-dioxopeperazine halves with the following functional groups: 1,3-disubstituted indole, 2,3-dihydro-2,3,3-trisubstituted indole, three N-methyls, two sp3 methylenes, a 1,2-ethanediamino, a carbomethyl, and two hexasubstituted dioxopiperazines (Table 1). The two fragment ions m/z 156 and m/z 231 in the fragmentation pattern of compound 1 also indicated the presence of two hexasubstituted dioxopiperazines (see Fig. S3 in the supplementary information). The 2,3,3-trisubstituted dihydroindolyl and 1,3-disubstituted indolyl groups were further supported by fragment ions m/z 115, 116, 129 [m/z 115+CH2]+, and 130 [m/z 116+CH2]+ in the fragmentation pattern of 1 (see Fig. S3 in the supplementary information), and the 1,3-disubstituted indolyl group was also supported by UV spectral data [275 nm (sh) (log ε 4.8), 288 (sh) (5.6), 295 (6.6)].6,11) The NMR data for one half of the compound 1 molecule showed a very similar pattern to those of chetomin,5,6) except that the 3-hydroxymethyl group [δΗ 4.29, 4.35 (each 1H, d, J=12.6 Hz, H2-13), 60.6 (CH2, C-13)] in chetomin (2) is replaced with a carbomethyl group [δC 206.5 (qC, C-12), 2.08 (3H, s, H3-13), 30.7 (CH3, C-13)] in 1, and a disulfide bridge [δC 74.8 (qC, C-3), 73.6 (qC, C-11a)] in chetomin (2) is replaced with a monosulfide bridge [δC 77.2 (qC, C-3), 73.7 (qC, C-11a)] in 1 (Table 1). This unit is further supported by the following mass fragments, m/z 646 [M+H−COCH3]+ and m/z 284 in the FAB-tandem MS (MS/MS) spectrum, and m/z 414 and 253 [285−S]+ in the EI-MS spectrum (see Figs. S1, S3, and S6 in the supplementary information). Detailed analysis of the NMR one and two dimensional (1D and 2D) and MS data of the other half revealed signals ascribable to indolyl-CH2–, hexasubstituted dioxopiperazine, 1,2-ethanediamino, and monochloro groups (Table 1). These functional groups are connected to each other according to COSY and HMBC data (Table 1) and the connection is further supported by the EI-MS fragments, m/z 116, 130, 173 [231−HNCH2CH2NH]+, 181 [231−CH3Cl]+, 182 [231+H−CH3Cl]+, 195 [231−HCl]+, 196 [231+H−HCl]+, 231, and 245 (see Fig. S3 in the supplementary information), and FAB-collision induced dissociation (CID) MS/MS spectra: m/z 400 [M+H−COCH3]+, 284, 245, 232 [245+H−CH2]+, and 183 [232+H−CH3Cl]+ (see Fig. S6 in the supplementary information). The key HMBC correlations from H3-15′ to C-3′ and from H-16′ and H2-17′ to C-3′ were also critical in establishing the position of the chlorine attached at C-3′ (Table 1). The ions at m/z 231, 195 [m/z 231−HCl]+, 183 [m/z 232+H−CH3Cl]+, and 181 [m/z 231−CH3Cl]+ also indicated that chlorine is located at C-3′ (see Figs. S3 and S6 in the supplementary information).
| Position | Cristazine (1) | ||
|---|---|---|---|
| δH (mult, J in Hz) | δC (mult) | HMBC (H to C) | |
| 1 | 165.3, qCc) | ||
| 3 | 77.2, qCe) | ||
| 4 | 165.4, qCc) | ||
| 5 | 6.14, d (1.6) | 80.2, CH | 6a, 10a, 10b |
| 6 | 7.32, br s | 5, 6a, 10a, 10b | |
| 6a | 149.1, qC | ||
| 7 | 6.73, d (7.5) | 110.3, CH | 9, 10a |
| 8 | 7.14, dd (7.5, 7.5) | 130.5, CH | 6a, 10 |
| 9 | 6.71, dd (7.5, 7.0) | 118.4, CH | 7, 10a |
| 10 | 7.37, d (7.0) | 124.6, CH | 6a, 8, 10b |
| 10a | 127.2, qC | ||
| 10b | 73.5, qCd) | ||
| 11 | 3.64, d (15.1) | 42.0, CH2 | 5, 10a, 10b |
| 3.99, d (15.1) | 1, 10a, 10b | ||
| 11a | 73.7, qCd) | ||
| 12 | 206.5, qC | ||
| 13 | 2.08, s | 30.7, CH3 | 3, 12 |
| 14 | 2.92, s | 27.9, CH3 | 1, 3 |
| 1′ | 165.2, qCc) | ||
| 3′ | 75.9, qC | ||
| 4′ | 161.0, qC | ||
| 6′ | 77.0, qCe) | ||
| 7′ | 3.81, AB q (15.5) | 26.5, CH2 | 1′, 6′, 8′, 9′, 14′a |
| 8′ | 107.8, qC | ||
| 9′ | 7.31, s | 127.0, CH | 7′, 8′, 10′a, 14′a |
| 10′a | 133.8, qC | ||
| 11′ | 7.48, d (8.0) | 111.4, CH | 13′, 14′a |
| 12′ | 7.15, dd (8.5, 8.0) | 122.1, CH | 10′a, 14′ |
| 13′ | 7.12, dd (8.5, 7.5) | 119.8, CH | 11′, 14′a |
| 14′ | 7.81, d (7.5) | 119.6, CH | 8′, 10′a, 12′ |
| 14′a | 129.9, qC | ||
| 15′ | 3.11, sb) | 27.8, CH3f) | 1′, 3′ |
| 16′ | 5.89, t (5.5) | 3′, 17′ | |
| 17′ | 4.23, dt (12.5, 5.5) | 59.2, CH2 | 3′ |
| 18′ | 4.31, dt (12.5, 5.5) | 58.7, CH2 | 6′ |
| 19′ | 5.94, t (5.5) | 6′, 18′ | |
| 20′ | 3.10, sb) | 27.7, CH3f) | 6′ |
a) Recorded in DMSO-d6 at 400 MHz (1H) and 100 MHz (13C). b–f) Interchangeable.
Therefore, the planar structure of cristazine represents a new class of dioxopiperazine alkaloid, shown as 1. The multiplet patterns and the nuclear Overhauser effect spectroscopy (NOESY) data for the α-protons are often analyzed to assign the relative configuration of diketopiperazine rings.12) But the α-carbons of diketopiperazine moieties in compound 1 are all quaternary, and attempts to secure the relative stereochemistry about the five chiral centers in 1 were precluded by unavailable NOESY data, except for between H-5a and Ha-11 and between H-7′ and H-19′. Thus, the stereochemistry illustrated for 1 is based on optical rotation and circular dichroism (CD) experiments, which established that 1 and chetomin (2)5,6) have identical stereochemistry at all asymmetric centers. Cristazine (1) showed the same sign of optical rotation ([α]D +220, CHCl3) as chetomin ([α]D +278, CHCl3)5) and a positive first Cotton effect at 303 (Δε +12.2)] and a negative Cotton effect at 263 nm (Δε −3.7), which indicates that 1 has the same stereochemistry as chetomin5,6) and pestalazine B.12)
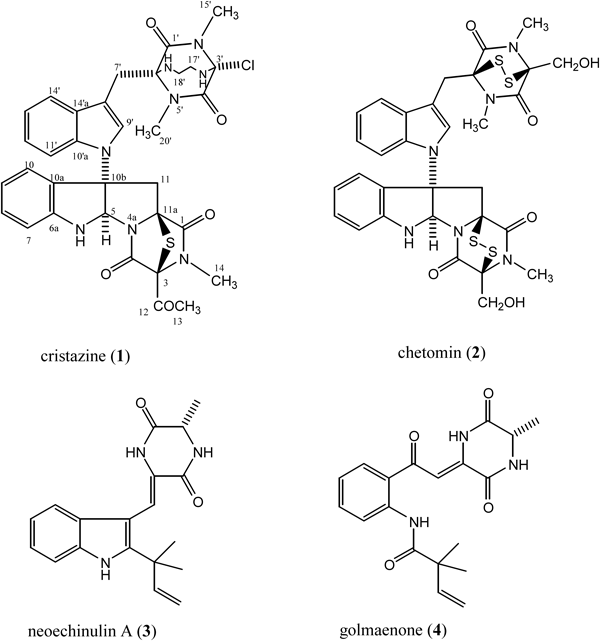
The dioxopiperazines 2–4 were also isolated in this study, and were identified by inspecting their NMR spectra and comparing those data with values in the literature.5–8)
Compounds 1–4 displayed potent radical-scavenging activities against 2,2-diphenyl-1-picrylhydrazyl (DPPH), with IC50 values of 19, 15, 24, and 20 µM, respectively, which were similar to that of the positive control, ascorbic acid (IC50, 20 µM). We comprehensively examined compound 1 for cytotoxic activity against the human cervical carcinoma (HeLa) cell line with a cell viability assay,13) and the IC50 value for 1 was 0.5 µM. Details of the inhibitory activity of 1 against HeLa cells is interesting, and will be reported in due course.
The instruments used to obtain the physicochemical data were the same as those described in our previous paper.14)
Fungal Isolation and CultureThe fungal strain, Chaetomium cristatum, was isolated from the sediments of marine mudflat collected at Suncheon Bay, Korea, and identified based on 18S ribosomal RNA (rRNA) analyses (SolGent Co., Ltd., Daejeon, Korea), identity of 97%. A voucher specimen is deposited at Pukyong National University with the code MSA295. The fungus was cultured in 3.0 L round flasks (20×1 L) for three weeks (static) at 29°C in the medium consisted of soytone (0.1%), soluble starch (1.0%), and seawater (100%).
Extraction and IsolationThe mycelium and broth were separated by filtration using cheeesecloth. The mycelium was extracted with CH2Cl2–MeOH (1 : 1) to afford mycelium extract (520 mg), which was subjected to Si gel flash chromatography. Elution was performed with n-hexane–EtOAc (stepwise, 0–100% EtOAc) to yield six fractions. Fraction 3, which exhibited radical scavenging activity against DPPH, was separated by medium-pressure liquid chromatography (MPLC) (octadecyl silica (ODS)) using a H2O–MeOH gradient elution to afford crude compounds 1 and 2. These were further purified by HPLC (Gemini C18, 4.6×250 mm, 5 µm, 1 mL/min) utilizing a 30 min gradient program of 50% to 100% MeOH in H2O to furnish 1 (8.5 mg) and 2 (2.5 mg). Compounds 3 (3.2 mg) and 4 (4.0 mg) were isolated from fraction 5, which also showed radical scavenging activity, by the same chromatographic method above.
Cristazine (1): colorless solid. [α]D20 +220 (c=0.3, CHCl3). mp 249–250°C. UV λmax (MeOH) nm (log ε) 275 (sh) (4.8), 288 (sh) (5.6), 295 (6.6). CD λmax (MeOH) nm (Δε) 303 (+12.2), 269 (−3.7), 238 (+31.9), 220 (+7.7), 202 (+26.9). IR (KBr) cm−1: 3484, 3355, 1681, 1608, 1457, 1345, 1203, 1062, 744. 1H- and 13C-NMR: see Table 1. Low resolution (LR)-EI-MS m/z : 414 ([M+H−COCH3]+−232) (rel. int. 15), 299 (46), 285 (13), 283 (34), 281 [299−CH3]+ (18), 269 (16), 266 [281−CH3]+ (12), 253 [285−S]+ (14), 245 (16), 238 [285−S−CH3]+ (4), 231 (1), 216 [231−CH3]+ (2), 196 [231+H−HCl]+ (1), 195 [231−HCl]+ (2), 182 [231+H−CH3Cl]+ (49), 181 [231−CH3Cl]+ (6), 173 [231−HNCH2CH2NH]+ (4), 170 (17), 169 (24), 166 [196−(CH3)2]+ (1), 156 (20), 155 [170−CH3]+ (100), 130 [361−231]+ (66), 129 (15), 128 (21), 116 [Indolyl]+ (4), 115 [Indolyl]+ (6), 114 (7), 101 (27), 77 (19), 56 (18), 43 [COCH3]+ (3), 42 [43-H]+ (23). LR-FAB-MS m/z : 646 [M+H−COCH3]+, 613 [646−SH]+. High resolution (HR)-EI-MS m/z : 43.0186 [COCH3]+ (Calcd for C2H3O, 43.0184) (+5.0 ppm/+0.2 mmu). HR-FAB-MS m/z: 646.1868 [M+H−COCH3]+ (Calcd for C31H31ClN8O4S, 646.1878) (−1.4 ppm/−0.9 mmu).
Chetomin (2), neoechinulin A (3), and golmaenone (4): spectroscopic data were virtually identical to those reported in the literature.5–8)
Radical Scavenging Assays15)Biological activity was measured exactly as described in our previous paper.
Cell Viability Assay13)To estimate cell viability, HeLa cells were seeded (1×104 cells) in 96-well plates in 100 µL of 10% fetal bovine serum-Dulbecco’s modified Eagle’s medium (FBS-DMEM) and cultured for 24 h at 37°C in a 5% CO2 incubator. After incubation, the cells were treated with 0, 12, 24, 36, 48, 60 and 80 µM of compounds and incubated for additional 24 h at 37°C in a 5% CO2 incubator. After treatment, 10 µL of EZ-Cytox cell viability assay solution WST-1® (Daeil Lab Service, Jong-ro, Korea) was added to each well, and the cells were then incubated for 3 h at 37°C in a 5% CO2 incubator. Absorbance was measured at 460 nm with enzyme-linked immunosorbent assay (ELISA) reader (Molecular Devices, Silicon Valley, CA, U.S.A.).
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2014R1A1A2053912).
The authors declare no conflict of interest.
The online version of this article contains supplementary materials. 1H-, 13C-NMR, and HMBC spectra of 1 (in DMSO-d6), LR-EI-MS, HR-EI-MS, LR-FAB-MS, HR-FAB-MS, FAB-CID MS (MS/MS) spectra of 1, fragmentation patterns of 1 in the EI-MS and FAB-MS. These materials are available free of charge via the Internet at http://pubs.acs.org.