2016 Volume 64 Issue 7 Pages 952-956
2016 Volume 64 Issue 7 Pages 952-956
Oryza sativa L. (rice) is an important staple crop across the world. In the previous study, we identified 36 specialized (secondary) metabolites including 28 flavonoids. In the present study, a metabolome analysis using liquid chromatography-mass spectrometry was conducted on the leaf, bran, and brown and polished rice grains to better understand the distribution of these metabolites. Principal component analysis using the metabolome data clearly characterized the accumulation patterns of the metabolites. Flavonoids, e.g., tricin, tricin 7-O-rutinoside, and tricin 7-O-β-D-glucopyranoside, were mainly present in the leaf and bran but not in the polished grain. In addition, anti-inflammatory and anti-oxidant activity of the metabolites were assayed in vitro. Tricin 4′-O-(erythro-β-guaiacylglyceryl)ether and isoscoparin 2″-O-(6‴-(E)-feruloyl)-glucopyranoside showed the strongest activity for inhibiting nitric oxide (NO) production and 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical-scavenging, respectively.
Plants produce a wide range of chemically and biologically diverse specialized (secondary) metabolites. These metabolites play an important role in human health because they have nutritional and medicinal aspects.1) Rice (Oryza sativa L.) is one of the most important staple crops and provides more than a fifth of the total calories consumed across the world.2)
The major nutrient of rice is starch; minor nutrients include protein, lipid, fiber, vitamins, and minerals.3) Rice accumulates bioactive specialized metabolites,4) such as phenylpropanoids,5–9) quinolone and pyridine alkaloids,10,11) terpenoids,12–14) and flavonoids (quercetin, kaempferol, tricin, and their glycosides).15–20) Such metabolites exhibit properties beneficial to humans, including anti-tumor,21) anti-inflammatory,22) and anti-oxidant activities.8) To improve the quality of rice plants, the organ-specific metabolic networks of primary and specialized metabolites were recently investigated using metabolite profiling based on liquid chromatography (LC)-MS.19,23–28) Metabolome-genome-wide association studies on rice have been increasingly attracting attention.25,29)
There are two primary types of rice grown in the world (i.e., the Indica and Japonica varieties). The Habataki cultivar is an Indica variety with higher yields, while the Koshihikari cultivar is a Japonica variety of better quality. In the previous study, 36 metabolites were isolated from the leaves of the Habataki cultivar.20) It is important to investigate organ specificity and benefits of the metabolites for application studies to improve the quality of rice plants. Here, to better understand the organ specificity of the metabolites and their benefits to humans, we performed the metabolite profiling of the leaf, bran, and brown and polished rice grains, using liquid chromatography-quadrupole time-of-flight (LC-QTOF)-MS. In addition, nitric oxide (NO) assay and 1,1-diphenyl-2-picrylhydrazyl (DPPH) assay using the metabolites were carried out for investigating their anti-inflammatory and anti-oxidant activities, respectively.
We have identified the distribution of the metabolites in the leaf, bran, and brown and polished rice grains, using LC-QTOF-MS. In this profiling, we used authentic standard compounds isolated from rice (Fig. 1): tricin (1), tricin 7-O-β-D-glucopyranoside (2), tricin 5-O-β-D-glucopyranoside (3), tricin 7-O-rutinoside (4), tricin 7-O-neohesperidoside (5), tricin 7-O-(2″-O-β-D-glucopyranosyl)-β-D-glucuronopyranoside (6), tricin 7-O-(6″-O-malonyl)-β-D-glucopyranoside (7), tricin 7-O-(6″-(E)-sinapoyl)-β-D-glucopyranoside (8), tricin 4′-O-(threo-β-syringylglyceryl)ether 7″-O-β-D-glucopyranoside (9), tricin 4′-O-(erythro-β-guaiacylglyceryl)ether (10), tricin 4′-O-(threo-β-guaiacylglyceryl)ether (11), tricin 4′-O-(erythro-β-guaiacylglyceryl)ether 7-O-β-D-glucopyranoside (12), tricin 4′-O-(threo-β-guaiacylglyceryl)ether 7-O-β-D-glucopyranoside (13), tricin 4′-O-(erythro-β-guaiacylglyceryl)ether 7″-O-β-D-glucopyranoside (14), tricin 4′-O-(threo-β-guaiacylglyceryl)ether 7″-O-β-D-glucopyranoside (15), tricin 4′-O-(erythro-β-guaiacylglyceryl)ether 9″-O-β-D-glucopyranoside (16), tricin 4′-O-(threo-β-4-hydroxyphenylglyceryl)ether (17), syringetin 3-O-β-D-glucopyranoside (18), syringetin 3-O-rutinoside (19), apigenin 6-C-α-L-arabinosyl-8-C-β-L-arabinoside (20), chrysoeriol 6-C-α-L-arabinosyl-8-C-β-L-arabinoside (21), swertisin (22), isoorientin 7,3′-dimethyl ether (23), luteolin 6-C-(2″-O-β-D-glucopyranosyl)-α-L-arabinoside (24), isoscoparin 2″-O-(6‴-(E)-feruloyl)-glucopyranoside (25), isoscoparin 2″-O-(6‴-(E)-p-coumaroyl)-glucopyranoside (26), isovitexin 2″-O-(6‴-(E)-feruloyl)-glucopyranoside (27), isovitexin 2″-O-(6‴-(E)-p-coumaroyl)-glucopyranoside (28), 1,3-O-diferuloylglycerol (29),1-O-feruloyl-β-D-glucose (30), 1-O-sinapoyl-β-D-glucose (31), 3-O-p-coumaroylquinic acid (32), 3-O-feruloylquinic acid (33), salicylic acid 2-O-β-D-glucopyranoside (34), kynurenic acid (35), and lycoperodine-1 (36).20)

Glc, β-D-glucopyranosyl; Rut, rutinosyl; Neo, neohesperidosyl; GluA, glucuronopyranosyl; Ara, arabinosyl. This figure is cited from the open access article, Yang et al., Metabolomics, 10, 543–555 (2014).
As shown in Fig. 2, among the 36 standard metabolites, 22 were detected in the samples. Compounds 1, 2, 4, 20, 30, and 36 were detected in the brown grain or bran. It has been reported that pigmented rice has a higher quantity of phenolic acids than non-pigmented rice.6) The amount of phenolic acids in brown rice is higher than that found in polished rice.6,8) Flavonoids accumulate in the rice bran of black rice cultivars but not in popular white rice cultivars.16,18,22) This profiling showed that the level of C-glycosylflavonoids (20–23) is higher in Habataki rice than that in Nipponbare. It has been reported that the Indica rice variety exhibits higher C-glycosylflavonoid production than the Japonica variety.19,24,25,29)
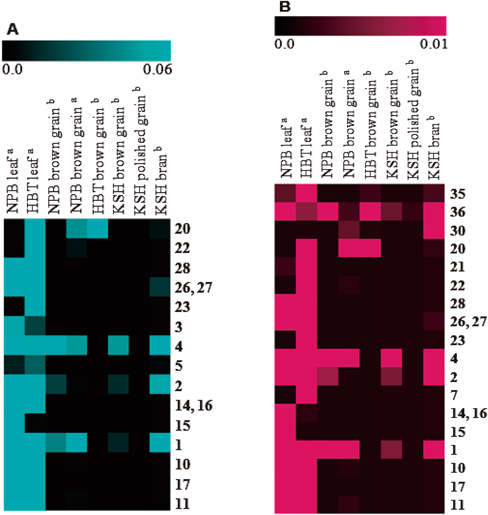
(A) Result of LC-QTOF-MS analysis in negative ion mode. (B) Result of LC-QTOF-MS analysis in positive ion mode. Color bar indicates relative signal intensity. NPB, Nipponbare; HBT, Habataki; KSH, Koshihikari. a, Sample grown in the laboratory, b: sample purchased from a market.
Principal component analysis (PCA) using the metabolome data characterized the accumulation patterns of the metabolites in the samples and showed the organ specificity of the metabolites (Fig. 3). Tricin (1), flavonolignans (10, 11) and flavonoid C-glycoside (20) contributed to the differences among the three groups. The signal intensities of these metabolites were higher in the leaf, suggesting that these components can be used as potential chemical markers to distinguish leaf from others.

The analysis was performed with three independent biological replicates for each sample. (A) Score plot. (B) Loading plot. Tricin (1), tricin 4′-O-(erythro-β-guaiacylglyceryl)ether (10), tricin 4′-O-(threo-β-guaiacylglyceryl)ether (11), apigenin 6-C-α-L-arabinosyl-8-C-β-L-arabinoside (20).
Compounds 1–4, 10, 12, 13, 25–27, 32, and 36 were examined with respect to their inhibition of lipopolysaccharide (LPS) and interferon-γ (IFN-γ) stimulated NO production in RAW264.7 cells and their DPPH free-radical-scavenging activity (Table 1). Quercetin was used as the positive control in both assay and showed IC50 values of 17.1 µM for the inhibiting NO production and 18.8 µM for DPPH radical scavenging. NO is known to play a pivotal role in inflammation and immune reactions. However, excessive production of NO may cause tissue damage and is thought to be involved in various types of inflammation.30) Compounds 3 and 10 showed moderate inhibition of NO production, having IC50 values of 75.1 and 32.6 µM, respectively. Compound 3 showed stronger inhibitory effect than compound 2, suggesting that the location of the sugar at C-5 was important for the anti-inflammatory activity. Compound 10 has been reported to have anti-inflammatory activity.22) It has been reported that compound 10 showed non inhibitory activity of NO production, while compound 1 exhibited inhibitory activity at the concentration of 40 µg/mL.31) In the DPPH radical-scavenging assay, compounds 25 and 36 showed weak antioxidant activity with IC50 values of 69.9 and 91.9 µM, respectively, as compared to that of quercetin. The other metabolites showed no significant anti-inflammatory or anti-oxidant activities at concentration of 100 µM.
| Compounds | IC50 (µM) | |
|---|---|---|
| NO | DPPH | |
| Tricin (1) | >100 | >100 |
| Tricin 7-O-β-D-glucopyranoside (2) | >100 | >100 |
| Tricin 5-O-β-D-glucopyranoside (3) | 75.1±2.1 | >100 |
| Tricin 7-O-rutinoside (4) | >100 | >100 |
| Tricin 4′-O-(erythro-β-guaiacylglyceryl)ether (10) | 32.6±0.6 | >100 |
| Tricin 4′-O-(erythro-β-guaiacylglyceryl)ether 7-O-β-D-glucopyranoside (12) | >100 | >100 |
| Tricin 4′-O-(threo-β-guaiacylglyceryl)ether 7-O-β-D-glucopyranoside (13) | >100 | >100 |
| Isoscoparin 2″-O-(6‴-(E)-feruloyl)-glucopyranoside (25) | >100 | 69.9±1.5 |
| Isoscoparin 2″-O-(6‴'-(E)-p-coumaroyl)-glucopyranoside (26) | >100 | >100 |
| Isovitexin 2″-O-(6‴-(E)-feruloyl)-glucopyranoside (27) | >100 | >100 |
| 3-O-p-Coumaroylquinic acid (32) | >100 | >100 |
| Lycoperodine-1 (36) | >100 | 91.9±6.8 |
| Quercetin (control compound) | 17.1±1.5 | 18.8±2.9 |
Values are shown as the mean±S.E.M. (n=3).
Metabolome analysis has become a useful technology to evaluate the relationship between the metabolites of plants and identify their biomarkers. The present study showed not only the accumulation patterns of metabolites in rice but also the potential health-promoting activities of the rice specialized metabolites. The biosynthetic mechanisms of the specialized metabolites are largely unknown in rice. The current study provides the basis to identify specific biosynthetic genes involved in metabolites production in phytochemical genomics study32,33) with a view to improve the quality of rice plants.
The leaf samples (Habataki and Nipponbare cultivars) and the brown rice sample (Nipponbare cultivar) were harvested from the rice plants grown in our laboratory, as previously reported.20) The other brown rice samples (Nipponbare and Koshihikari cultivars), along with the bran and polished rice, were purchased from a local market. All samples were immediately frozen, lyophilized, ground, and stored at −80°C until required.
LC-QTOF-MS AnalysisThe extraction27) and LC-QTOF-MS analysis20) was conducted as previously described.
Data AnalysisData acquisition and processing were performed using the MassLynx 4.1 and Progenesis CoMet (Nonlinear Dynamics, Durham, NC, U.S.A.) software. Heat map visualization was performed using MeV (www.tm4.org).
NO Production Inhibition and DPPH Radical-Scavenging AssayThe inhibition of NO production in the macrophage-like cell line, RAW264.7 activated with LPS and IFN-γ, was conducted as previously described.34) The DPPH assay procedure followed a previous method with slight modifications.34) Acetic acid buffer 0.5 M (15 µL), ethanol (145 µL), and sample (0.4 µL) were mixed with 0.5 mM DPPH (40 µL). The resulting solution was thoroughly mixed, and kept at room temperature in dark. After 30 min, the absorbance was measured at 520 nm. The DPPH radical-scavenging activity was determined by comparing the absorbance of the test sample with that of the control (100%), which contained only DPPH and the solvent.
We thank Mr. Kouji Takano for the technical assistance. This research was supported, in part, by the Japan Science and Technology Agency (JST), Strategic International Collaborative Research Program (SICORP), and by the Japan Advanced Plant Science Research Network.
The authors declare no conflict of interest.