2016 Volume 64 Issue 7 Pages 979-981
2016 Volume 64 Issue 7 Pages 979-981
The biogenetic origins of amphidinin A (1) and amphidinolide P (2) were investigated by feeding experiments with 13C-labeled acetates. 13C-NMR data of 13C-enriched samples revealed that the all carbons of 1 and 2 were derived from acetates. The polyketide chain of 1 was formed from one triketide chain, two diketide chains, and three unusual isolated C1 units derived from C-2 of cleaved acetates, while the polyketide chain of 2 was formed from one pentaketide chain, two acetate units, and three unusual isolated C1 units derived from C-2 of cleaved acetates. The all branched C1 units of 1 and 2 were derived from C-2 of cleaved acetates.
Marine dinoflagellate has been recognized as the organism produce novel secondary metabolites with intriguing structures and significant biological activities.1) In our continuing search for bioactive metabolites from marine dinoflagellates, we have isolated a series of macrolides, amphidinolides, and linear polyketides from symbiotic dinoflagellates belonging to the genus Amphidinium.2–7) The carbon skeletons of dinoflagellate polyketides are unique and unavailable from other organisms, since they might be generated by unexplained one-carbon extension machinery in addition to the ordinary polyketide biosynthesis.8) In our continuing biosynthetic study of dinoflagellate polyketides,9–16) we have recently re-isolated amphidinin A (1)17,18) and amphidinolide P (2)19,20) from the cultured marine dinoflagellate Amphidinium sp. (2012-7-4A strain) which were isolated from an acoel flatworm Amphiscolops sp. In this study, we have investigated the biogenetic origins of 1 and 2 by feeding experiments with [1-13C], [2-13C], and [1,2-13C2] sodium acetates to the culture of dinoflagellate Amphidinium sp. (2012-7-4A).
The [1-13C], [2-13C2], and [1,2-13C2] acetate labeled amphidinin A (1) and amphidinolide P (2) were purified by column chromatographies from the culture medium of dinoflagellate Amphidinium sp. (2012-7-4A) obtained by feeding experiments with corresponding 13C-labeled sodium acetates. The 13C incorporation ratios of [1-13C] and [2-13C] acetate labeled 1 and 2 were shown in Tables 1 and 2, respectively.
| Position | δC | Incorporation ratioa) | Assignmentb) | |
|---|---|---|---|---|
| [1-13C]Acetate | [2-13C]Acetate | |||
| 1 | 67.1 | 1.10 | 1.58 | m |
| 2 | 73.0 | 1.13 | 0.87 | c |
| 3 | 39.8 | 1.71 | 2.24 | m |
| 4 | 69.1 | 1.14 | 0.87 | c |
| 5 | 44.6 | 1.77 | 2.17 | m |
| 6 | 35.2 | 0.92 | 1.23 | m |
| 7 | 151.1 | 1.10 | 0.80 | c |
| 8 | 50.4 | 1.03 | 1.46 | m |
| 9 | 83.0 | 1.41 | 0.79 | c |
| 10 | 47.4 | 1.32 | 1.79 | m |
| 11 | 36.2 | 1.26 | 1.47 | m |
| 12 | 83.2 | 1.50 | 1.00 | c |
| 13 | 129.7 | 0.70 | 1.28 | m |
| 14 | 132.0 | 0.94 | 0.83 | c |
| 15 | 41.1 | 1.52 | 1.70 | m |
| 16 | 144.3 | 0.95 | 0.64 | c |
| 17 | 111.5 | 1.10 | 1.23 | m |
| 18 | 22.5 | 2.59 | 2.75 | m |
| 19 | 15.2 | 3.50 | 4.19 | m |
| 20 | 25.5 | 1.92 | 2.38 | m |
| 21 | 112.0 | 1.00 | 1.35 | m |
| 22 | 23.9 | 1.70 | 2.53 | m |
a) The 13C incorporation ratios were calculated based on the increase of the intensity of each signal on 13C-NMR spectra compared with corresponding signal on those of natural sample, assuming the intensity of C-21 and C-12 as 1.00 for [1-13C] and [2-13C]acetate labeled amphidinin A (1). b) The “c” and “m” denote the carbon derived from C-1 and C-2 of acetate, respectively.
| Position | δC | Incorporation ratioa) | Assignmentb) | |
|---|---|---|---|---|
| [1-13C]Acetate | [2-13C]Acetate | |||
| 1 | 172.4 | 1.39 | 1.00 | c |
| 2 | 45.01 | 1.37 | 1.62 | m |
| 3 | 99.2 | 0.85 | 1.09 | m |
| 4 | 45.2 | 1.11 | 1.52 | m |
| 5 | 143.7 | 1.08 | 1.00 | c |
| 6 | 39.4 | 1.30 | 1.76 | m |
| 7 | 73.5 | 1.32 | 0.96 | c |
| 8 | 62.8 | 1.29 | 1.62 | m |
| 9 | 58.2 | 1.61 | 1.26 | c |
| 10 | 36.4 | 1.47 | 1.87 | m |
| 11 | 142.3 | 1.20 | 0.79 | c |
| 12 | 133.6 | 1.00 | 1.23 | m |
| 13 | 129.1 | 1.19 | 1.07 | c |
| 14 | 78.5 | 0.95 | 1.24 | m |
| 15 | 45.04 | 1.09 | 1.40 | m |
| 16 | 146.5 | 1.15 | 0.88 | c |
| 17 | 112.3 | 1.18 | 1.45 | m |
| 18 | 16.1 | 3.62 | 4.10 | m |
| 19 | 110.0 | 1.02 | 1.42 | m |
| 20 | 11.8 | 3.11 | 3.83 | m |
| 21 | 19.5 | 2.81 | 2.90 | m |
| 22 | 118.1 | 1.14 | 1.30 | m |
a) The 13C incorporation ratios were calculated based on the increase of the intensity of each signal on 13C-NMR spectra compared with corresponding signal on those of natural sample, assuming the intensity of C-12 and C-1 as 1.00 for [1-13C] and [2-13C]acetate labeled amphidinolide P (2). b) The “c” and “m” denote the carbon derived from C-1 and C-2 of acetate, respectively.
Though all carbons were enriched in some measure by 13C labeled carbon dioxides generated by the metabolism of 13C labeled acetates,8) the 13C-NMR spectrum of amphidinin A (1) labeled with [1-13C] sodium acetate showed significant enrichments of 7 carbons (C-2, C-4, C-7, C-9, C-12, C-14, C-16). While significant enrichments of 15 carbons (C-1, C-3, C-5, C-6, C-8, C-10, C-11, C-13, C-15, C-17, C-18, C-19, C-20, C-21, C-22) were observed on the 13C-NMR spectrum of 1 labeled with [2-13C] sodium acetate. These results indicated that all the 22 carbons of 1 were derived from acetates (Fig. 1). The analysis of INADEQUATE spectrum of 1 labeled with [1,2-13C2] sodium acetate indicated that 7 C2 units (C-2/C-3, C-4/C-5, C-7/C-8, C-9/C-10, C-12/C-13, C-14/C-15, C-16/C-18) were come from intact acetates. Whereas the analysis implied that 3 C1 units (C-1, C-6, C-11) in the polyketide chain and 5 branched C1 units (C-17, C-19. C-20, C-21, C-22) were derived from C-2 of cleaved acetates. The origin of the terminal carbon of 1 (C-1) was not C-1 of acetate but C-2 of cleaved acetate. This incorporation pattern could be found in other dinoflagellate polyketides.8) Especially, this is the common feature of linear dinoflagellate polyketides.21–23)

The similar analysis revealed that all the 22 carbons of amphidinolide P (2) were originated from acetates (Fig. 2). In detail, the origins of 7 carbons (C-1, C-5, C-7, C-9, C-11, C-13, C-16) and 15 carbons (C-2, C-3, C-4, C-6, C-8, C-10, C-12, C-14, C-15, C-17, C-18, C-19, C-20, C-21, C-22) were C-1 and C-2 of acetate, respectively. The 7 C2 units (C-1/C-2, C-5/C-6, C-7/C-8, C-9/C-10, C-11/C-12, C-13/C-14, C-16/C-21) of them were derived from intact acetates. By contrast, the 3 C1 units (C-3, C-4, C-15) in the polyketide chain and 5 branched C1 units (C-17, C-18, C-19, C-20, C-22) were come from C-2 of cleaved acetates.
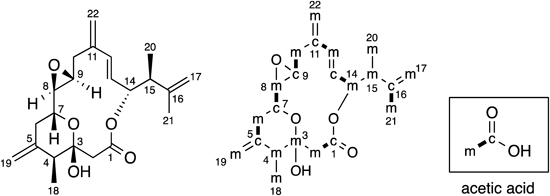
The incorporation of C1 unit derived from C-2 of cleaved acetate is the characteristic feature of dinoflagellate polyketides. The all branched C1 units, attaching to the C-1 of intact acetate or the C-2 of cleaved acetate in polyketide chain, were derived from C-2 of cleaved acetate. The direct experimental evidence has not been found yet, however, the incorporation patterns obtained from many feeding experiments indicated the possible involvement of the 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase (HCS) cassette to the addition of this kind of branched C1 unit.8,24) Though the mechanism of unusual dinoflagellate polyketide chain extension was unknown, C1 unit come from C-2 of cleaved acetate in polyketide chain might exist as carbonyl carbon at the early stage. In fact, almost all C1 units originated from C-2 of cleaved acetate in polyketide chain existed as modified carbon (carbonyl, methylated, hydroxylated, or olefinic). The methylenes at C-10 of amphidinolide T1 and C-17 of amphidinolide Y were the rare case for the carbon originated from C-2 of cleaved acetate.13,15) This hypothesis does not contradict the idea that the addition of branched C1 unit to C-2 of cleaved acetate was also done by HCS cassette.
1H-NMR spectra were recorded on a JEOL JNM-ECX500 spectrometer. 13C-NMR spectra were recorded on a Bruker DRX-600 spectrometer using 2.5 mm micro cells (Shigemi Co., Ltd., Japan) for C6D6. The 7.20 ppm resonances of residual C6D5H was used as internal references for 1H-NMR spectra. The 128 ppm resonances of C6D6 was used as internal references for 13C-NMR spectra.
Cultivation and IsolationThe dinoflagellates Amphidinium sp. (2012-7-4A) were separated from a flatworm Amphiscolops sp., which was collected at Ishigaki island, Okinawa, Japan. The dinoflagellates were cultured at 25°C under 16 h light/8 h dark schedule in 4× Provasoli’s enriched seawater (PES) medium (500 mL) with 2.5 mL of antibiotic–antimycotic mixed stock solution (Nacalai Tesque Inc. (Japan); Antibiotic–Antimycotic Mixed Stock Solution, 100×, Stabilized, Penicillin 10000 units/mL, Streptomycin 10000 µg/mL, Amphotericin B 25 µg/mL). The supernatant was aspirated at seventh day after inoculation, and flesh 4× PES medium (500 mL), antibiotic–antimycotic mixed stock solution (2.5 mL), and [1-13C], [2-13C], and [1,2-13C2] sodium acetate (0.5 mM) were added. After additional cultivation for 3 d, the supernatant was aspirated, passed through a filter paper, and subjected to a porous polymer gel column (Diaion HP-20, Mitsubishi Chemical Co., Japan, ϕ8.5×35.0 cm). The column was washed with H2O (4 L) and the adsorbed materials were eluted with MeOH (4 L), which were concentrated in vacuo. The MeOH eluted materials, obtained from 250 L (500 mL×20) of culture, were partitioned between n-hexane (500 mL×3) and H2O (500 mL) to afford n-hexane soluble materials. The n-hexane soluble materials were concentrated in vacuo and fractionated by a silica gel column (Wakogel C-300, Wako Pure Chemical Industries, Ltd. (Japan), ϕ1.0×8 cm; eluent, n-hexane–EtOAc, 5 : 1 to 0 : 1, then EtOAc–MeOH, 100 : 0 to 0 : 100). The fractions containing [1-13C], [2-13C], and [1,2-13C2]acetate labeled amphidinin A (1) were separated by C18 HPLC (Luna 5u C18(2), Phenomenex Inc. (Japan), ϕ10×250 mm; eluent, MeCN–H2O, 55 : 45; flow rate, 2.5 mL/min; UV detection at 210 nm) to afford [1-13C], [2-13C], and [1,2-13C2]acetate labeled amphidinin A (1, 0.6, 0.4, 1.0 mg, respectively). While the fractions containing [1-13C], [2-13C], and [1,2-13C2]acetate labeled amphidinolide P were separated by C18 HPLC (Luna 5u C18(2), Phenomenex Inc., ϕ10×250 mm; eluent, MeCN–H2O, 75 : 25; flow rate, 2.5 mL/min; UV detection at 220 nm) to afford [1-13C], [2-13C], and [1,2-13C2]acetate labeled amphidinolide P (2, 0.5, 0.2, 0.3 mg, respectively).
We thank Dr. E. Fukushi, Graduate School of Agriculture, Hokkaido University, for measurements of INADEQUATE spectrum. This work was partially supported by the Naito Foundation, the Takeda Science Foundation, MEXT-Supported Program for the Strategic Research Foundation at Private Universities (2013–2017), and JSPS KAKENHI Grant Number 23108502.
The authors declare no conflict of interest.
The online version of this article contains supplementary materials.