2017 年 65 巻 3 号 p. 236-247
2017 年 65 巻 3 号 p. 236-247
Certain pyridazine containing compounds 2a–f, 3a, b, 4a, b, 5a, b, 6a and b were synthesized and characterized by spectroscopic means and elemental analysis. All the synthesized compounds were screened for their cytotoxic activity in vitro on colon cancer cell line (HCT-116) and breast cancer cell line (MCF-7). In addition, the antitumor activity of the synthesized compounds was tested in vivo against Ehrlich’s ascites carcinoma (EAC) solid tumor grown in mice. The in vitro vascular endothelial growth factor receptor (VEGFR) enzyme inhibition assay was carried out for the most active compounds at a single dose of 10 µM. The obtained results revealed that compound 5b, which showed potent cytotoxic activity against HCT-116 also, exhibited the highest inhibition in the VEGFR kinase assay (92.2%).
Among all diseases that affect humanity, cancer ranks high as a major killer.1) Surgery and radiation are used to treat cancer that is confined locally, whereas drug therapy is essential to kill cancer cells that have spread to distant sites in the body. Drug treatment mainly involved cytotoxic chemotherapy that kills all rapidly dividing cells, both tumor and normal.2) The optimum goal is to find a treatment modality that specifically kills malignant cells and causes little or no side effects. Therefore, targeted therapy was developed to target key elements that play a role in tumor development and tumor growth. In addition, tyrosine kinase inhibitors (TKI) were developed to block intracellular signaling pathways in tumor cells, leading to deregulation of key cell functions as proliferation and differentiation.3) The receptor tyrosine kinase family includes receptors for many growth factors such as epidermal growth factor (EGF), platelet-derived growth factor (PDGF) and vascular endothelial growth factor (VEGF).4) Accordingly, modern drug research has become focused on signal transduction therapy.5) Inhibition of VEGF receptor-2 (VEGFR-2) signaling pathway was proved to provide an antiangiogenic effect on human cancer.6)
The first TKI to be used clinically is imatinib (Gleevec®)7) (Fig. 1). TKI mimic ATP for binding with ATP binding domain of VEGF and VEGFRs leading to blockade of intracellular signaling, thus preventing the activation of downstream effectors involved in tumor progression.8) It was reported that treatment with imatinib was associated with the inhibition of VEGF expression and secretion in vitro in neuroblastoma cells. The decrease in VEGF expression was associated with suppression of in vivo tumor growth.9) In the current work, imatinib was chosen as a standard drug for the in vitro cytotoxic assay and the in vivo anticancer assay in mice.7,9,10)

From another point of view, pyridazine moiety has been successfully incorporated in several potent anticancer compounds I11) (Fig. 2). Furthermore, some pyridazine containing compounds II were reported to have an inhibitory activity against VEGFR-212) (Fig. 2). The encouraging anticancer activity observed by pyridazine derivatives prompted us to investigate a new analogues.

Accordingly, the design of our compounds was based on the replacement of the carboxylic group in the previously prepared pyridazine derivatives11) III by different groups as H, CH3, F, 2,6-Cl2, Br, NHCOCH3, NHCOCH2CH2Cl, NHCSNHCH2CH3 or NHCSNHCH2CH=CH2. In addition, we replace benzenecarboxylic acid moiety by 2-chloro-3-pyridyl moiety to study the effect of the new compounds on the cytotoxic activity (Fig. 3).
Additionally, pyrimidine ring in imatinib was replaced by pyridazine ring in our compounds which is connected with the aryl system via NH group as in imatinib. According to the literature survey it was obvious that some pyridazine containing compounds possess promising cytotoxic activity.1,11,13) In addition, pyridazine is considered as bioisostere of pyrimidine. Thus, it was interesting to investigate the effect of replacement of pyrimidine by pyridazine on the cytotoxic activity of the new compounds.

Furthermore, sulfonamide group was incorporated replacing piperazine moiety hoping that the prepared compounds will attain superior activity (Fig. 3).
The synthetic pathways for target compounds 2a–f is outlined in Chart 1. The intermediate 1 was prepared from the commercially available 3,6-dichloropyridazine and sulfaguanidine, according to a reported procedure.11) Compounds 2a–f were obtained by reacting 1 with different aromatic amines. 1H-NMR spectrum of compound 2b showed a singlet signal at δ 2.35 ppm integrated for three protons corresponding to the CH3 of 4-methylphenyl. New pyridazine derivatives 3a, b, 4a, b, 5a, b, 6a and b were synthesized according to Chart 2. Adopting nucleophilic substitution reaction of 1 with either meta or para phenylenediamine gave the relevant compounds 3a and b. Reaction of 3a with either acetyl chloride or 3-chloropropionyl chloride afforded 4a and b, respectively. IR spectra of compounds 4a and b proved to be useful in tracing the appearance of C=O stretching at 1647 and 1676 cm−1 of O=C–R, respectively. 1H-NMR spectrum of compound 4a revealed the presence of a singlet signal at δ 2.02 ppm integrated for three protons due to CH3 of O=C–CH3. Also, 1H-NMR spectrum of compound 4b demonstrated the presence of two triplets at δ 2.83 and 3.89 ppm integrated for four protons corresponding to CH2CH2Cl. Reacting compounds 3a and b with different isothiocyanates yielded the corresponding compounds 5a, b, 6a and b. 1H-NMR spectra of compounds 5a and 6a showed the presence of a triplet at δ 1.12 ppm and a quartet at δ 3.50 and 3.47 ppm, respectively assigned for the ethyl group. Both the analytical and spectral data (IR, 1H-NMR, 13C-NMR and MS) of the newly synthesized compounds were in full agreement with the proposed structures.14)

Reagents and conditions: i)  OH/reflux 4 h, ii) ArNH2/CH3CH2CH2CH2OH/reflux 15h.
OH/reflux 4 h, ii) ArNH2/CH3CH2CH2CH2OH/reflux 15h.

Reagents and conditions: i)  /C2H5OH/reflux 12 h, ii) RCOCl/ C2H5OH/reflux 9 h. iii) RNCS/C2H5OH/reflux 10 h.
/C2H5OH/reflux 12 h, ii) RCOCl/ C2H5OH/reflux 9 h. iii) RNCS/C2H5OH/reflux 10 h.
The antitumor activity of compounds 2a–f, 4a, b, 5a, b, 6a and b was evaluated in vitro using colon cancer cell line (HCT-116) and breast cancer cell line (MCF-7) applying Sulforhodamine B stain (SRB) colorimetric assay. For comparison purpose, the cytotoxic activity of imatinib (Fig. 1), a standard antitumor drug for treatment of gastrointestinal tract tumors7) and breast tumors15) that acts as VEGFR inhibitor,16) was evaluated under the same condition. The IC50 values are shown in Table 1. The results are represented graphically in Figs. 4 and 5. The analysis of data in Table 1 showed that all the tested compounds produced significant cytotoxic activity. Compound 5b was especially more potent than imatinib with IC50 equivalent to 30.3 µM, where it bears allylthiourea group. Compounds 4a, b, 6a and b showed comparable cytotoxicity to imatinib. Moreover, compounds 2a, b, d, e, f and 5a were less active than imatinib, and compound 2c was the least active among all the tested compounds against colon cancer cell line (HCT-116). These biological results revealed that methyl group in compound 2b gave better activity than unsubstituted phenyl in compound 2a. Moreover, substituent as bromide in compound 2f, increases the potency, while substituent, as fluoride in compound 2c, decreases the potency. The presence of two chlorine atoms as in compound 2e exhibited moderate activity. Furthermore, 2-chloro-3-pyridyl moiety in compound 2d showed comparable activity compared to the substituted phenyl group in compounds 2a–e and f. On the other hand, the presence of bulky substituent, such as, allyl group in compound 5b, increases the activity.


| Compound number | (IC50)a,b) in µM against (HCT-116) | (IC50)a,b) in µM against (MCF-7) |
|---|---|---|
| 2a | 51.8 | 53.4 |
| 2b | 44.2 | 111.2 |
| 2c | 66.7 | 64.5 |
| 2d | 49.8 | 48.4 |
| 2e | 50.6 | 36.2 |
| 2f | 48.4 | 34.8 |
| 4a | 35.1 | 25.6 |
| 4b | 39.0 | 21.2 |
| 5a | 40.3 | 25.5 |
| 5b | 30.3 | 27.5 |
| 6a | 33.7 | 23.6 |
| 6b | 33.5 | 25.3 |
| Imatinibc) | 34.3 | 6.0 |
a) IC50: dose of the compound which inhibit tumor cell proliferation by 50%. b) Values are means of three experiments. c) Used as positive control.
On the other hand, analysis of data in Table 1 revealed that compound 4b (IC50 21.2 µM) was the most active among the synthesized compounds against breast cancer cell line (MCF-7). This compound contains aminophenyl-3-chloropropanamide moiety at position 6 of the pyridazine ring, while other compounds were found to exhibit poor activity. The in vitro cytotoxic activity on breast cancer cell line (MCF-7) indicated that methyl group in compound 2b showed lower potency than unsubstituted phenyl in compound 2a. In addition, substituent, as bromide in 2f, relatively increases the activity, while fluoride in compound 2c, decreases the activity. Moreover, the presence of two chlorine atoms in compound 2e possess good cytotoxic activity. On the other hand, 2-chloro-3-pyridyl moiety in compound 2d exhibited moderate activity. Furthermore, no significant change in potency resulted from changing the substituents in compounds 4a, 5a, b, 6a and b.
In Vivo AssayThe final target compounds 2a–f, 4a, b, 5a, b, 6a and b were screened for their antitumor activity against Ehrlich’s ascites carcinoma (EAC) solid tumors grown in mice. Among them, compounds 2a, b, e, f, 4a, b, 5b and 6b displayed comparable activity to that produced by imatinib. Importantly, the antitumor activity produced by compounds 2e, 4b and 5b was greater than that produced by imatinib at p<0.05 (Fig. 6). Where the tumor weights were 12, 10 and 33 mg, respectively compared to 390 mg in imatinib. These three most active compounds were selected for further elucidation through testing their anti-angiogenic effect.

Data are expressed as the mean±S.E.M. and analyzed using one-way ANOVA followed by Bonferroni’s post-hoc test. * Compared to the tumor control group at p<0.05. $ Compared to imatinib at p<0.05.
The histological criteria used for scoring of solid tumors in particular, necrosis area, mitotic picture and giant cell in solid tumors taken from the experimental groups were expressed in Fig. 7. The scores for necrosis area, mitotic picture and giant cell as well as the total score in solid tumors taken from mice treated with imatinib or compounds 2e, 4b and 5b were illustrated in Table 2. The antitumor activity of these synthesized compounds was expressed through decrease in the total score compared to the tumor control.
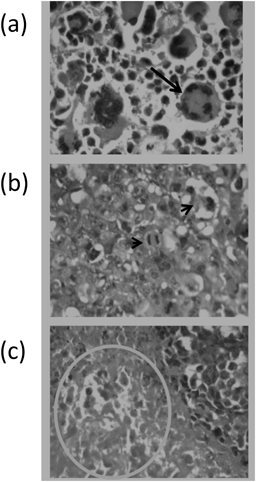
a) Tumor giant cell (solid arrow); b) Mitotic picture (arrow heads); c) Necrosis area (outlined by the circle). Ehrlich’s carcinoma cell line was inoculated intradermally in the back of female mice and dissected at the end of the experiment, fixed in paraformaldehyde solution and stained with H&E. Photomicrographs are captured at 40×magnification.
| Groups | Necrosis area | Mitotic picture | Giant cell | Mean total score |
|---|---|---|---|---|
| Tumor control | 3.000±0.000 | 3.000±0.000 | 3.000±0.000 | 9.000±0.000 |
| Imatinib | 0.910±0.170a) | 1.000±0.000a) | 1.000±0.000a) | 2.920±0.140a) |
| 2e | 1.200±0.200a) | 1.200±0.200a) | 2.400±0.240a,b) | 4.900±0.530a,b) |
| 4b | 2.300±0.190a) | 1.800±0.200a,b) | 2.100±0.210a,b) | 6.300±0.300a,b) |
| 5b | 1.460±0.200b) | 2.280±0.210b) | 2.290±0.170a,b) | 6.120±0.350a,b) |
Solid tumors stained with hematoxylin and eosin were scored as (0) absent, (1) low or weak, (2) mild to moderate and (3) high or frequent. Then, the total score was calculated and averaged for each group. Data are expressed as the mean±S.E.M. and analyzed using one-way ANOVA followed by Bonferroni’s post-hoc test. a) Compared to the vehicle group at p<0.05. b) Compared to imatinib at p<0.05.
Statistical analysis revealed that serum VEGF level was lower in the experimental groups treated with imatinib or compounds 2e, 4b and 5b in comparison to the tumor control group (540.000±48.300, 435.800±53.800, 591.000±78.010 and 596.000±111.000 vs. 1266.830±82.660) at p<0.05 (Fig. 8); there was no statistical difference among the last four treated groups. Immunohistochemical staining of VEGFR in solid tumor sections highlighted a decrease in immunostaining in imatinib group by 30% and a decrease in groups treated with compounds 2e, 4b and 5b by 35, 10 and 40%, respectively compared to the tumor control group (Fig. 9). The in vivo antitumor activity against EAC solid tumor grown in mice showed that the presence of two chlorine atoms, as in compound 2e, showed significant activity in comparison to tumor control and imatinib. In addition, the presence of bulky groups, as in compounds 4b and 5b, exhibit comparable antitumor activity to that of imatinib.

VEGF: vascular endothelial growth factor. Data are expressed as the mean±S.E.M. and analyzed using one-way ANOVA followed by Bonferroni’s post-hoc test. * Compared to the tumor control group at p<0.05.

Data are expressed as the mean±S.E.M. and analyzed using one-way ANOVA followed by Bonferroni’s post-hoc test. * Compared to the tumor control group at p<0.05.
To assess the VEGFR kinase inhibitory activity of the newly synthesized compounds, as a suggested mechanism of action, the in vitro kinase inhibitory activity against MCF-7 and HCT-116 was evaluated for the most active compounds, as cytotoxic agents using VEGFR kinase inhibitory activity assay by enzyme-linked immunosorbent assay (ELISA) (results summarized in Table 3). The inhibitory activity was given as percentage inhibition at concentration of 10 µM. The tested compounds exhibited significant VEGFR inhibitory activity with percentage inhibition (89.6–92.0%) against MCF-7 where they were more potent than imatinib (78.7%). On the other hand, compounds 5b and 6a exhibited significant cytotoxic activity against HCT-116 with IC50 (30.3, 33.7, respectively), showing high VEGFR kinase inhibitory activity (92.2, 91.5%, respectively), comparable to that produced by imatinib (92.1%).
| Compound number | % Inhibition (MCF-7) | % Inhibition (HCT-116) |
|---|---|---|
| 4b | 89.6% | ND |
| 5a | 91.2% | ND |
| 5b | 92.0% | 92.2% |
| 6a | 91.7% | 91.5% |
| Imatinib | 78.7% | 92.1% |
ND: not determined.
In an effort to elucidate the anti-angiogenic mechanism by which the synthesized compounds can induce anticancer activity, molecular docking study was performed. It is used to predict the binding modes and orientation of the synthesized compounds at the active site of the ATP binding site of VEGFR. The coordinates of the VEGF structure were obtained from the crystal structure of VEGFR, which forms a complex with its inhibitor (PDB code 3CJG).17) This revealed the hydrogen bond interactions between N1 of the pyrimidine and NH of aniline Cys 917, two of the methoxy groups with asparagine Asn 921 and N of the pyrazole ring with Glu 883 (Fig. 10). The docking algorithm was validated via re-docking of the co-crystallized ligand in the VEGFR active site where the docking procedure was able to retrieve the co-crystallized pose with root-mean-square deviation (RMSD) value of 1.81 Å. Docking of the investigated compounds revealed that pyridazine derivatives 4b, 5b and 6a were capable of occupying the ATP binding site of the VEGFR kinase, while maintaining the essential key interactions. In binding model, compound 4b is nicely bound to VEGFR with its NH of aniline group, and N1 of the pyridazine ring with Cys 917 and S=O binds with Glu 883, which are the same amino acids that interact with the lead compound. Additionally, C=NH interacts with Lys 866, and NH2 of the guanidine group binds with the backbone of Asp 1044, which increases the potency. Moreover, S=O of compound 5b form interaction with Glu 883, C=NH binds to Lys 866 and NH2 of guanidine group interacts with Asp 1044. On the other hand, the binding model of compound 6a showed that it is bound to VEGFR through NH of aniline group and N1 of the pyridazine ring with Cys 917 and S=O binds with Glu 883, which are the same amino acids that interact with the lead compound. Additionally, C=NH interacts with Lys 866 and NH2, NH of the guanidine group bind with the backbone of Asp 1044, which increases the potency of 6a (as it exhibited significant VEGFR kinase inhibitory activity with percentage inhibition 91.7% against MCF-7 and 91.5% against HCT-116).
According to these docking results, guanidine moiety made two additional hydrogen bonds with Lys 866 and Asp 1044 which indicates better binding with the active site of ATP binding site of VEGFR. One of the best-scored poses of compounds 4b, 5b and 6a is presented, respectively, in Figs. 11–13.




The present study reports the synthesis and anticancer activity of certain pyridazine containing compounds 2a–f, 4a, b, 5a, b, 6a and b targeting VEGFR kinase. The results revealed that compounds 4a, b, 6a and b showed cytotoxic activity comparable to the reference drug imatinib on colon cancer cell line (HCT-116), while compound 5b revealed cytotoxic activity with IC50 value lower than that of imatinib, with percentage enzyme inhibition 92.2% comparable to that produced by imatinib (92.1%). Other compounds were found to exhibit poor activity. Moreover, compound 4b was the most active one against breast cancer cell line (MCF-7) with IC50 equivalent to 21.2 µM. On the other hand, the antitumor activity produced by compounds 2e, 4b and 5b was greater than that produced by imatinib, when tested in vivo against EAC where percentage enzyme inhibition of 4b and 5b (89.6, 92.0%, respectively) were more significant than that of imatinib (78.7%).
Molecular docking study of 4b and 6a on VEGFR kinase active site revealed their ability to form the essential hydrogen bond interaction with Cys 917 and Glu 883 which are the same amino acids that interact with the lead compound, with binding energy scores equal to −20.3715, −21.9495 and −16.0372 kcal/mol, respectively. Additionally, C=NH interacts with Lys 866. Moreover, NH2 of the guanidine group in compound 4b binds with the backbone of Asp 1044, while NH and NH2 of guanidine in compound 6a bind with Asp 1044, which increases the potency of the tested compounds.
Finally, molecular docking models of compounds 4b and 6a indicated that they bind to the ATP binding site of VEGFR forming the essential hydrogen bond interaction. This result coincides with the percentage inhibition of the VEGFR, where 4b showed percentage inhibition 89.6% against MCF-7, and 6a exhibited percentage inhibition 91.7% against MCF-7 and 91.5% against HCT-116. The above results may suggest that the designed compounds act as VEGFR inhibitors.
Melting points (mp) were determined on Griffin apparatus, and the values given are uncorrected. IR spectra were determined on Shimadzu IR 435 spectrophotometer (KBr, cm−1). 1H-NMR spectra were carried out using Varian Mercury-300 (300 or 400 MHz) Spectrophotometer using tetramethylsilane (TMS) as internal standard. Chemical shift values are recorded in ppm on δ scale, Micro Analytical Center, Cairo University, Egypt and Micro Analytical Unit, Faculty of Pharmacy, Cairo University, Egypt. Coupling patterns are described as follows: s, singlet, d, doublet, t, triplet, m, multiplet. J describes a coupling constant. Mass spectra were recorded on a GCMP-QP1000 EX Mass spectrometer, Micro Analytical Center, Cairo University, Egypt. Elemental analyses were carried out at the Micro Analytical Center, Al-Azhar University, Egypt. Progress of the reactions was monitored using TLC sheets pre-coated with UV fluorescent silica gel Merck 60F 254, using toluene–ethanol (4.5 : 0.5) and were visualized using UV lamp. All chemicals were obtained from Aldrich, Fluka, or Merck chemicals. N-Carbamimidoyl-4-[(6-chloropyridazin-3-yl)amino]benzensulfonamide (1) was prepared as reported.11)
General Procedure for the Preparation of 2a–fAn equimolar amount of 1 (3.265 g, 0.01 mol) and the appropriate amine (0.01 mol) in 20 mL n-butanol was heated under reflux for 15 h. The reaction mixture was cooled, and the separated solid was filtered, dried, and recrystallized from ethanol.
N-Carbamimidoyl-4-[(6-phenylaminopyridazin-3-yl)amino]benzensulfonamide (2a)Yield: 1.63 g (42.6%); mp: 190–192°C; 1H-NMR (dimethyl sulfoxide (DMSO)-d6, 300 MHz) δ: 6.70 (2H, s, D2O exchangeable), 7.36 (1H, d, J=8.40 Hz), 7.11–7.76 (9H, m), 7.57 (1H, d, J=8.40 Hz), 9.80 (1H, s, D2O exchangeable), 10.03 (1H, s, D2O exchangeable). 13C-NMR (DMSO-d6, 100 MHz) δ: 113.43, 121.17, 122.35, 126.77, 127.81, 129.77, 130.07, 131.46, 134.71, 151.13, 158.10. IR (KBr) cm−1: 3435, 3338, 3267, 3184, 3132, 1330 and 1138. MS m/z: 385.00 [M+2]+, 384 [M+1]+, 383.00 (M+), 291.00, 234.00. Anal. Calcd for C17H17N7O2S (383.43): C, 53.25; H, 4.47; N, 25.57; Found: C, 53.31; H, 4.49; N, 25.71.
N-Carbamimidoyl-4-[(6-(4-methylphenyl)aminopyridazin-3-yl)amino]benzensulfonamide (2b)Yield: 0.78 g (19.6%); mp: 130–132°C; 1H-NMR (DMSO-d6, 400 MHz,) δ: 2.35 (3H, s), 6.59 (2H, d, J=8.56 Hz), 6.73 (2H, s, D2O exchangeable), 7.22 (2H, d, J=8.04 Hz), 7.43 (2H, d, J=8.60 Hz), 7.56 (1H, s, D2O exchangeable), 7.73 (1H, d, J=8.76 Hz), 7.80 (2H, d, J=8.72 Hz), 7.87 (1H, d, J=8.76 Hz), 9.55 (1H, s, D2O exchangeable), 9.76 (1H, s, D2O exchangeable). 13C-NMR (DMSO-d6, 100 MHz) δ: 20.98, 113.13, 116.66, 117.91, 121.41, 127.25, 127.74, 130.19, 133.39, 137.40, 150.51, 158.50. IR (KBr) cm−1: 3452, 3433, 3371, 3332, 3215, 2850–2900, 1332 and 1130. MS m/z: 397.00 (M+), 233.00. Anal. Calcd for C18H19N7O2S (397.45): C, 54.39; H, 4.82; N, 24.67; Found: C, 54.47; H, 4.91; N, 24.83.
N-Carbamimidoyl-4-[(6-(2-fluorophenyl)aminopyridazin-3-yl)amino]benzensulfonamide (2c)Yield: 1.56 g (39.0%); mp: 196–198°C; 1H-NMR (DMSO-d6, 300 MHz,) δ: 5.65 (2H, s, D2O exchangeable), 6.53 (2H, d, J=6.60 Hz), 6.54 (2H, d, J=6.60 Hz), 6.55 (1H, d, J=8.40 Hz), 6.45–7.79 (4H, m), 7.36 (1H, d, J=8.40 Hz), 9.80 (1H, s, D2O exchangeable). 13C-NMR (DMSO-d6, 100 MHz) δ: 110.76, 112.79, 115.68, 118.21, 121.29, 124.60, 127.23, 127.70, 130.03, 131.28, 147.99, 151.83, 158.22. IR (KBr) cm−1: 3454, 3433, 3371, 3331, 3219, 1303 and 1128. MS m/z: 401.00 (M+), 400.00 [M−1]+, 291.00 233.00. Anal. Calcd for C17H16FN7O2S (401.42): C, 50.87; H, 4.02; N, 24.43; Found: C, 50.96; H, 4.08; N, 24.55.
N-Carbamimidoyl-4-[(6-(2-chloropyridin-3-ylamino)pyridazin-3-yl)amino]benzensulfonamide (2d)Yield: 1.74 g (41.7%); mp: 184–186°C; 1H-NMR (DMSO-d6, 300 MHz) δ: 5.65 (2H, s, D2O exchangeable), 6.53 (2H, d, J=6.60 Hz), 6.54 (1H, d, J=8.40 Hz), 6.55–7.79 (3H, m), 7.37 (2H, d, J=6.60 Hz), 7.38 (1H, d, J=8.40 Hz), 9.80 (1H, s, D2O exchangeable). 13C-NMR (DMSO-d6, 100 MHz) δ: 112.81, 118.20, 121.34, 127.22, 127.71, 130.05, 131.28, 137.56, 143.15, 148.58, 151.80, 158.22. IR (KBr) cm−1: 3489, 3435, 3417, 3398, 3342, 3223, 1309 and 1134. MS m/z: 420.10 [M+2]+, 418.10 (M+), 416.10 [M−2]+, 290.10. Anal. Calcd for C16H15ClN8O2S (418.86): C, 45.88; H, 3.61; N, 26.75; Found: C, 45.96; H, 3.67; N, 27.02.
N-Carbamimidoyl-4-[(6-(2, 6-dichlorophenyl)aminopyridazin-3-yl)amino]benzensulfonamide (2e)Yield: 0.65 g (14.4%); mp: 260–262°C; 1H-NMR (DMSO-d6, 400 MHz) δ: 6.53 (2H, d, J=8.64 Hz), 6.66 (2H, s, D2O exchangeable), 7.22–7.69 (3H, m), 7.37 (2H, d, J=8.64 Hz), 7.70 (1H, d, J=8.80 Hz), 7.81 (1H, d, J=8.80 Hz), 9.49 (1H, s, D2O exchangeable), 9.84 (1H, s, D2O exchangeable). 13C-NMR (DMSO-d6, 100 MHz) δ: 112.89, 118.20, 121.33, 127.22, 127.72, 130.05, 137.56, 143.14, 148.58, 157.01, 158.48.IR (KBr) cm−1: 3431, 3329, 3234, 3203, 3169, 1253 and 1138. MS m/z: 456.00 [M+4]+, 454.00 [M+2]+, 452.00 (M+), 292.00, 233.00. Anal. Calcd for C17H15Cl2N7O2S (452.32): C, 45.14; H, 3.34; N, 21.68; Found: C, 45.16; H, 3.37; N, 21.74.
N-Carbamimidoyl-4-[(6-2-bromophenylaminopyridazin-3-yl)amino]benzensulfonamide (2f)Yield: 0.60 g (13.0%); mp: >300°C; 1H-NMR (DMSO-d6, 300 MHz,) δ: 6.57 (2H, s, D2O exchangeable), 6.65–7.82 (8H, m), 7.26 (1H, d, J=9.30 Hz), 7.62 (1H, d, J=9.30 Hz), 7.83 (1H, s, D2O exchangeable); 9.57 (1H, s, D2O exchangeable), 9.83 (1H, s, D2O exchangeable). 13C-NMR (DMSO-d6, 100 MHz) δ: 113.85, 118.20, 121.37, 127.22, 127.90, 131.56, 133.08, 135.23, 143.22, 148.58, 150.65, 157.91, 158.45. IR (KBr) cm−1: 3421, 3350, 3329, 3232, 3203, 3170, 1253 and 1138. MS m/z: 463.00 [M+2]+, 461.00 (M+), 460.00 [M−1]+, 234.00. Anal. Calcd for C17H16BrN7O2S (462.32): C, 44.16; H, 3.49; N, 21.21; Found: C, 44.20; H, 3.53; N, 21.29.
General Procedure for the Preparation of (3a, b)A mixture of 1 (3.265 g, 0.01 mol) and the appropriate phenylenediamine (1.08 g, 0.01 mol) in 30 mL absolute ethanol was heated under reflux for 12 h. The reaction mixture was concentrated under reduced pressure, cooled and the formed solid was filtered, dried and recrystallized from ethanol.
N-Carbamimidoyl-4-[(6-(3-aminophenylamino)pyridazin-3-yl)amino]benzensulfonamide (3a)Yield: 2.93 g (73.8%); mp: 130–132°C; 1H-NMR (DMSO-d6, 400 MHz,) δ: 6.64–7.81 (4H, m), 6.67 (1H, d, J=8.56 Hz), 6.79 (2H, s, D2O exchangeable), 6.91 (2H, d, J=7.08 Hz), 7.36 (2H, d, J=7.52 Hz), 7.46 (1H, d, J=8.60 Hz), 7.62 (1H, s, D2O exchangeable), 10.45 (1H, s, D2O exchangeable), 10.52 (1H, s, D2O exchangeable). 13C-NMR (DMSO-d6, 100 MHz) δ: 111.86, 113.58, 116.17, 127.27, 127.84, 130.50, 131.50, 133.07, 135.23, 150.32, 150.96, 158.07, 158.52. IR (KBr) cm−1: 3435, 3400, 3344, 3215, 1226 and 1130. MS m/z: 399.00 [M+1]+, 398.00 (M+), 292.00, 233.00. Anal. Calcd for C17H18N8O2S (398.44): C, 51.25; H, 4.55; N, 28.12; Found: C, 51.41; H, 4.59; N, 28.34.
N-Carbamimidoyl-4-[(6-(4-aminophenylamino)pyridazin-3-yl)amino]benzensulfonamide (3b)Yield: 3.48 g (87.6%); mp: 96–98°C; 1H-NMR (DMSO-d6, 300 MHz,) δ: 5.52 (2H, s, D2O exchangeable), 6.52 (2H, d, J=6.60 Hz), 6.53 (2H, d, J=6.60 Hz), 6.63 (1H, d, J=8.70 Hz), 7.28 (1H, d, J=8.70 Hz), 7.36 (2H, d, J=6.90 Hz), 7.37 (2H, d, J=6.90 Hz), 8.03 (1H,s, D2O exchangeable), 9.00 (1H, s, D2O exchangeable), 9.80 (1H, s, D2O exchangeable). 13C-NMR (DMSO-d6, 100 MHz) δ: 110.02, 112.92, 114.89, 116.45, 118.31, 127.72, 128.15, 131.32, 141.40, 151.69, 158.22. IR (KBr) cm−1: 3489, 3435, 3414, 3398, 3342, 3219, 1232 and 1132. MS m/z: 398.10 (M+), 397.10 [M−1]+, 291.10, 233.00. Anal. Calcd for C17H18N8O2S (398.44): C, 51.25; H, 4.55; N, 28.12; Found: C, 51.37; H, 4.58; N, 28.27.
General Procedure for the Preparation of (4a, b)An equimolar amount of 3a (3.98 g, 0.01 mol) and the selected acid chloride (0.01 mol) in 25 mL absolute ethanol was heated under reflux for 9 h. The reaction mixture was concentrated under reduced pressure and the formed solid was filtered dried and recrystallized from ethanol.
N-(3-(6-(4-(N-Carbamimidoylsulfamoyl)phenylamino)pyridazin-3-ylamino)phenyl)acetamide (4a)Yield: 0.59 g (13.4%); mp: 196–198°C; 1H-NMR (DMSO-d6, 300 MHz) δ: 2.02 (3H, s), 6.29 (2H, s, D2O exchangeable), 6.56 (1H, d, J=8.70 Hz), 6.62 (2H, d, J=6.90 Hz), 6.63 (2H, d, J=6.90 Hz), 6.66–7.74 (4H, m), 7.39 (1H, d, J=8.70 Hz), 10.20 (1H, s, D2O exchangeable). 13C-NMR (DMSO-d6, 100 MHz) δ: 40.96, 114.92, 117.15, 119.25, 123.01, 127.42, 128.45, 129.22, 131.17, 132.72, 136.49, 146.81, 147.95, 157.03, 158.68. IR (KBr) cm−1: 3442, 3394, 3342, 3309, 3224, 3209, 3140, 2922–2854, 1647, 1244 and 1136. MS m/z: 440.20 (M+), 439.20 [M−1]+, 291.10, 233.10. Anal. Calcd for C19H20N8O3S (440.48): C, 51.81; H, 4.58; N, 25.44; Found: C, 51.89; H, 4.56; N, 25.71.
N-(3-(6-(4-(N-Carbamimidoylsulfamoyl)phenylamino)pyridazin-3-ylamino)phenyl)-3-chloropropanamide (4b)Yield: 0.70 g (14.3%); mp: 150–152°C; 1H-NMR (DMSO-d6, 300 MHz,) δ: 2.83 (2H, t, J=6.30 Hz), 3.89 (2H, t, J=6.30 Hz), 6.33 (2H, s, D2O exchangeable), 6.27–7.95 (8H, m), 6.61 (1H, d, J=8.40 Hz), 7.43 (1H, d, J=8.70 Hz), 9.94 (1H, s, D2O exchangeable), 10.08 (1H, s, D2O exchangeable), 10.39 (1H, s, D2O exchangeable). 13C-NMR (DMSO-d6, 100 MHz) δ: 33.61, 55.09, 114.50, 117.23, 119.03, 121.86, 127.13, 127.35, 127.45, 128.01, 130.73, 135.43, 150.41, 157.74, 158.48, 169.74. IR (KBr) cm−1: 3446, 3342, 3315, 3251, 3201, 3151, 2962–2835, 1676, 1313 and 1136. MS m/z: 491.10 [M+3]+, 489.10 [M+1]+, 291.10, 233.00. Anal. Calcd for C20H21ClN8O3S (488.95): C, 49.13; H, 4.33; N, 22.92; Found: C, 49.24; H, 4.39; N, 23.10.
General Procedure for the Preparation of (5a, b)A solution of 3a (3.98 g, 0.01 mol) and the appropriate isothiocyanate (0.01 mol) in 25 mL absolute ethanol was heated under reflux for 10 h. The solution was concentrated under reduced pressure and the residue triturated with ether. The formed solid was filtered, dried and recrystallized from ethanol.
N-Carbamimidoyl-4-(6-(3-(3-ethylthiourea)phenylamino)pyridazin-3-ylamino)benzensulfonamide (5a)Yield: 4.24 g (87.4%); mp: 180–182°C; 1H-NMR (DMSO-d6, 400 MHz,) δ: 1.12 (3H, t, J=7.08 Hz), 3.50 (2H, q, J=7.20 Hz), 6.69 (2H, s, D2O exchangeable), 6.54–7.79 (8H, m), 7.55 (1H, d, J=8.64 Hz), 7.65 (1H, d, J=8.64 Hz), 8.08 (1H, s, D2O exchangeable), 9.58 (1H, s, D2O exchangeable), 9.81 (1H, s, D2O exchangeable). 13C-NMR (DMSO-d6, 100 MHz) δ: 14.45, 65.39, 113.73, 119.00, 121.77, 122.90, 126.64, 127.90, 129.38, 131.48, 139.37, 142.66, 150.81, 157.93, 158.51, 180.36. IR (KBr) cm−1: 3462, 3446, 3433, 3334, 3292, 3246, 3213, 3149, 3109, 2972–2929, 1338 and 1141. MS m/z: 484.90 [M−1]+, 291.90, 233.00. Anal. Calcd for C20H23N9O2S2 (485.59): C, 49.47; H, 4.77; N, 25.96; Found: C, 49.62; H, 4.82; N, 26.13.
N-Carbamimidoyl-4-(6-(3-(3-allylthiourea)phenylamino)pyridazin-3-ylamino)benzensulfonamide (5b)Yield: 2.12 g (42.6%); mp: 82–84°C; 1H-NMR (DMSO-d6, 300 MHz,) δ: 4.12 (2H, t), 5.10–5.34 (2H, m), 5.83–5.94 (1H, m), 6.67 (2H, s, D2O exchangeable), 6.52–7.80 (8H, m), 7.58 (1H, d, J=8.70 Hz), 7.65 (1H, d, J=8.70 Hz), 7.83 (1H, s, D2O exchangeable), 8.16 (1H, s, D2O exchangeable), 9.89 (1H, s, D2O exchangeable), 10.34 (1H, s, D2O exchangeable). 13C-NMR (DMSO-d6, 100 MHz) δ: 46.43, 112.92, 116.18, 116.41, 120.29, 121.82, 126.57, 127.72, 131.32, 134.90, 135.22, 139.55, 142.68, 151.69, 158.23, 158.56, 180.94. IR (KBr) cm−1: 3423, 3369, 3348, 3334, 3217, 2900–2850, 1328 and 1136. MS m/z: 499.00 [M+2]+, 498.00 [M+1]+, 497.00 (M+). Anal. Calcd for C21H23N9O2S2 (497.60): C, 50.69; H, 4.66; N, 25.33; Found: C, 50.78; H, 4.70; N, 25.47.
General Procedure for the Preparation of (6a, b)A mixture of 3b (3.98 g, 0.01 mol) and the appropriate isothiocyanate (0.01 mol) in 25 mL absolute ethanol was heated under reflux for 10 h. The reaction mixture was concentrated under reduced pressure and the residue triturated with ether. The formed solid was filtered, dried and recrystallized from ethanol.
N-Carbamimidoyl-4-(6-(4-(3-ethylthiourea)phenylamino)pyridazin-3-ylamino)benzensulfonamide (6a)Yield: 1.59 g (32.7%); mp: 90–92°C; 1H-NMR (DMSO-d6, 400 MHz) δ: 1.12 (3H, t, J=7.20 Hz), 3.47 (2H, q, J=6.36 Hz), 5.68 (1H, s, D2O exchangeable), 6.52 (2H, d, J=8.56 Hz), 6.68 (2H, s, D2O exchangeable), 7.37 (2H, d, J=8.56 Hz), 7.54 (1H, d, J=8.60 Hz), 7.65 (1H, d, J=8.60 Hz), 7.69 (2H, d, J=8.72 Hz), 7.80 (2H, d, J=8.72 Hz), 8.02 (1H, s, D2O exchangeable), 9.72 (1H, s, D2O exchangeable). 13C-NMR (DMSO-d6, 100 MHz) δ: 14.45, 41.82, 112.82, 118.21, 123.91, 126.64, 127.21, 127.71, 131.26, 139.47, 142.59, 151.82, 158.53, 180.43. IR (KBr) cm−1: 3429, 3334, 3230, 2900–2850, 1334 and 1134. MS m/z: 487.00 [M+2]+, 486.00 [M+1]+, 485.00 (M+), 232.00. Anal. Calcd for C20H23N9O2S2 (485.59): C, 49.47; H, 4.77; N, 25.96; Found: C, 49.55; H, 4.74; N, 26.13.
N-Carbamimidoyl-4-(6-(4-(3-allylthiourea)phenylamino)pyridazin-3-ylamino)benzensulfonamide (6b)Yield: 1.72 g (34.6%); mp: 104–106°C; 1H-NMR (DMSO-d6, 300 MHz) δ: 4.11 (2H, t), 5.07–5.22 (2H, m), 5.65 (2H, s, D2O exchangeable), 5.82–5.93 (1H, m), 6.52 (1H, d, J=8.40 Hz), 6.88 (2H, d),7.35 (2H, d), 7.36 (1H, d, J=8.40 Hz), 7.37 (2H, d), 7.38 (2H, d), 7.83 (1H, s, D2O exchangeable), 9.20 (1H, s, D2O exchangeable), 9.60 (1H, s, D2O exchangeable), 9.80 (1H, s, D2O exchangeable). 13C-NMR (DMSO-d6, 100 MHz) δ: 46.51, 112.82, 116.17, 116.43, 124.01, 126.60, 127.71, 131.28, 134.91, 135.29, 135.98, 151.81, 158.23, 158.53, 181.07. IR (KBr) cm−1: 3452, 3431, 3371, 3331, 3211, 3194, 2999–2856, 1301 and 1130. MS m/z: 498.00 [M+1]+, 497.00 (M+), 232.00. Anal. Calcd for C21H23N9O2S2 (497.60): C, 50.69; H, 4.66; N, 25.33; Found: C, 50.82; H, 4.70; N, 25.39.
Antitumor ActivityMaterials and MethodsIn Vitro AssayThe cytotoxic activity of the tested compounds was measured in vitro against human colon cancer cell line (HCT-116) and breast cancer cell line (MCF-7) in comparison to imatinib, applying Sulforhodamine B (SRB) stain following the method of Skehan et al.10) Human tumor cell lines (MCF-7, HCT-116) used in this study were obtained frozen in liquid nitrogen (−180°C) from the American Type Culture Collection (ATC C, MN, U.S.A.). The tumor cell lines were maintained at the National Cancer Institute, Cairo, Egypt, by serial sub-culturing. Cells were seeded in 96-well microtiter plates at a concentration of (5×104–105cells/well) in a fresh medium and left to attach to the plates for 24 h before treatment of the tested compounds. Test compounds were dissolved in DMSO, and diluted with saline to the appropriate volume. After 24 h, cells were incubated with the appropriate concentration ranges of drugs (0, 5, 12.5, 25 and 50 µg/mL), the wells were diluted to 200 µL with fresh medium and incubation was continued for 48 h at 37°C. Control cells were treated with vehicle alone. Four wells were used for each drug concentration. After 48 h incubation, the cells were fixed with 50 µL of cold 50% trichloroacetic acid for 1 h at 4°C, washed 5 times with distilled water and then stained for 30 min at room temperature with 50 µL 0.4% SRB dissolved in 1% acetic acid. The wells were then washed 4 times with 1% acetic acid. The plates were air dried and the dye was solubilized with 100 µL/well of 10 mM tris base (pH=10.5) for 5 min on a shaker (Orbital shaker OS 20, Boeco, Germany) at 1600 rpm. The optical density (O.D.) of each well was measured spectrophotometrically at 564 nm with an ELISA microplate reader (Meter tech. Σ 960, U.S.A.). The percentage of cell survival was calculated as follows: survival fraction=O.D. (treated cells)/ O.D. (control cells). The relation between surviving fraction and compound concentration was plotted to get the survival curve for tumor cell line after the specified time. The concentration required for 50% inhibition of cell viability (IC50) was calculated for each tested compound (Table 1).
In Vivo AssayInduction of EAC Solid Tumors in MiceFemale Swiss albino mice with original body weight of 20–26 g were used in the current experiment, and fixed housing conditions were maintained at a normal dark/light cycle, mice were housed in groups of twelve in polyethylene cages where food and water were provided ad libitum. The Research Ethics Committee at the Faculty of Pharmacy, Suez Canal University (license number 20148A2), approved the study of protocol. EAC is commonly employed as a solid form18,19) and easy to grow in suspension, when injected in the peritoneal cavity of female mice. Mice carrying EAC cell line were obtained from the Department of Tumor Biology at the National Cancer Institute (Cairo, Egypt). The viability of EAC cells was ensured employing Trypan blue dye exclusion method. The next step, EAC cells suspension was prepared in sterile saline solution to get a final working suspension; each 0.1 mL of which contained 2.5 million of EAC cells. At the beginning of the experiment, mice were shaved at their back, and inoculated with 0.1 mL of the EAC suspension.
Experimental DesignSeven days after inoculation with the tumor cells in all female mice, mice were randomly divided into fourteen groups, ten mice in each. The different treatments were started as follows; Group 1: mice treated daily with a 1 : 1 (v/v) mixture of DMSO/PEG400 (5 mL/kg/d, intraperitoneally (i.p.)). Group 2: mice treated daily with the standard chemotherapeutic agent, imatinib (10 mg/kg/d, i.p.). Groups 3–14: mice treated daily with compounds 2a–f, 4a, b, 5a, b, 6a and b (10 mg/kg/d, i.p.). All therapies continued for 10 d.
Dissection of Tumor Discs and Collection of Serum SamplesTumor MassOne day after the end of the experiment (day 18), mice were maintained under light ether anesthesia and blood samples were collected by cardiac puncture. Blood samples were maintained at room temperature for 30 min and centrifuged at 12000×g for 10 min. Then, sera were separated and stored at −20°C until used. An ELISA kit (Sun Red Biotechnology Company, Shanghai, China) was employed for estimating serum VEGF level according to the manufacturer’s protocol. The color intensity was measured at 450 nm using an automated ELISA reader (Europe S.A., Belgium).
Histopathological Examination of the Solid TumorsHenceforward, mice were sacrificed by cervical dislocation under anesthesia and solid tumors from each mouse were dissected. Then, tumors were weighed and fixed in 10% phosphate-buffered formalin. All paraffin-embedded sections were cut at 4 µm and stained with hematoxylin and eosin (H&E). Histopathological evaluation for solid tumor sections was focused in the extension of necrotic area, finding of typical mitotic picture and the presence of neoplastic giant cells. Each one of these findings was scored according to their frequency and intensity: (0) absent, (1) weak or low, (2) mild to moderate and (3) high or frequent. Then, the summation of each score was calculated, averaged and compared for the experimental groups. Histopathological examinations were evaluated blindly.20)
Immunohistochemistry and Image AnalysisAnother tissue section (4 µm) was used for immunohistochemical staining of VEGF receptors-2. Briefly, rabbit polyclonal VEGF receptor-2 antibodies were added to each slide and incubated overnight at 4°C. Then, biotinylated secondary antibodies were added followed by the enzyme conjugate and finally, 2 drops of 3,3-diaminobenzidine chromogen were added to each slide to visualize the immune reaction. Finally, cover slipping was performed, the slides were examined blindly under a light microscope and photomicrographs were captured. The photomicrographs were then assessed using the ImageJ 1.45 F image analysis system (National Institute of Health, U.S.A.) to elucidate the percent of area of immunostaining.
Statistical AnalysisData were collected, tabulated and presented as the mean±standard error of the mean (S.E.M.) One-way ANOVA followed by Bonferroni’s post-hoc test were used to analyze the difference between the experimental groups. All statistical tests were performed employing the statistical package for social science (Chicago, IL, U.S.A.), version 16. A p value <0.05 was considered to be statistically significant.21)
VEGFR Inhibitory Activity Assay by ELISAVEGFR-3 tyrosine (Tyr) kinase inhibitory activity was determined using Abcam’s VEGF Receptor 3 Human ELISA (Sanfrancisco, CA, U.S.A.). The procedure of the used kit was performed according to the manufacturer’s instructions.
Molecular Modeling StudyDocking StepsAll the docking study was done using Molecular Operating Environment (MOE 2008.10: Chemical Computing Group, Canada) as the computational software. In order to perform docking, preliminary steps were done.
Enzyme structures were checked for missing atoms, bonds and contacts. All water of crystallization was deleted away from the active site except the one involved in the interaction with the ligand. Hydrogens and partial charges were added to the system, using Protonate 3D application. The active site was isolated; recognition of the amino acids and the backbone was hidden. The interactions of the ligand with the amino acids of the active site were studied.
Docking of CompoundsThe compounds were built using MOE molecular builder and energy minimized by Merck Molecular Force Field (MMFF94x). Hydrogens and partial charges were added to the system, using Protonate 3D application. Compounds were grouped in databases, and docked into the active site, using the MOE Dock tool.
Conformational Analysis of CompoundsThe algorithm generated conformations from a single 3D conformation by conducting a systemic search. In this way, all combinations of angles were created for each compound. A collection of poses was generated from the pool of ligand conformations using Triangle Matcher placement method. Poses were generated by superposition of ligand atom triplets and triplets of points in the receptor-binding site in a systemic way. Poses generated by placement methodology were scored using an available method implemented in MOE, the London dG scoring function, which estimates the free energy of binding of the ligand from a given pose. The top30 poses for each ligand were output in a MOE database. Each resulting pose was then subjected to MMFF94x energy minimization. The minimized docking conformations were then rescored, using London dG scoring method.
The authors are grateful to all members of Cancer Biology Department, National Cancer Institute, Cairo, Egypt, for carrying out in vitro cytotoxic testing. Also we would like to express our deepest thanks to Dr. Mohamed Abdel Daim, Department of Pharmacology, Faculty of Veterinary Medicine, Suez Canal University, Egypt for helping in inoculating tumor cell line and Dr. Mohammed Kamal, Department of Pathology, Faculty of Medicine, Suez Canal University, Egypt for capturing photomicrographs. Furthermore, we would like to thank Dr. Esam Rashwan, manager of Confirmatory Diagnostic Unit, VACSERA, Egypt for performing the in vitro enzyme based inhibition assay.
The authors declare no conflict of interest.