2019 Volume 67 Issue 8 Pages 839-848
2019 Volume 67 Issue 8 Pages 839-848
Panacis Japonici Rhizoma (PJR) contains various kinds of saponins, which possesses extensive pharmacological activities, but studies of comprehensive analysis of its saponins were limited. Thus, ultra-fast liquid chromatography coupled with triple quadrupole-time of flight tandem mass spectrometry (UFLC-Triple TOF-MS/MS) and ultra-fast liquid chromatography coupled with triple quadrupole-linear ion trap tandem mass spectrometry (UFLC-QTRAP-MS/MS) methods were established for the qualitative and quantitative analysis of the saponins in PJR, separately. Fifty three saponins in PJR were identified by UFLC-Triple TOF-MS/MS method, 23 saponins of which were unequivocally identified by reference substances. In addition, fragmentation pathways of different types of saponins were preliminarily deduced by fragmentation behavior of 53 saponins. Furthermore, the simultaneous determination of the contents of 13 saponins in PJR samples harvested at different times were analyzed by UFLC-QTRAP-MS/MS method. Furthermore, the quality of the samples was evaluated by grey relational analysis. This study might be beneficial to the quality assessment and control of PJR. Meanwhile, it might provide the basic information for confirming its optimal harvested period.
As a member of Araliaceae Panax family, Panacis Japonici Rhizoma (PJR) is derived from the dried rhizome of Panax japonicus C. A. MEY,1) which has been considered as a highly valuable traditional Chinese medicine that was first documented in “Ben Cao Gang Mu Shi Yi” (published in 1765 A.D).2) In Chinese medicine, it has been used to scatter stasis detumescence, relieve pain and replenish deficiency for a long time.2) As the most abundant composition in PJR, saponins possess antioxidant, anti-inflammatory, anti-myocardial ischemia and other pharmacological activities.2–5) It is different from the chemical composition of P. ginseng, P. notoginseng and P. quinquefolium that the content of oleanane-type saponins was higher in PJR, whereas the content of dammarane-type saponins was relatively lower.5–9) Our previous study mostly focused on high content saponins and was in lack of a relatively comprehensive analysis of the saponins in PJR.5,10) Therefore, in support of the holistic quality assessment of PJR, it is necessary to develop a rational approach to analyze both the major and minor saponins in PJR.
Some techniques, such as NMR11,12) and liquid chromatography coupled with mass spectrometry (LC-MS),7) have been established for the identification and characterization of saponins in PJR. NMR can provide reliable and specific structural information for compounds, and is extensionally applicable in the isolation and identification of compounds. In comparison with NMR, LC-MS allows higher sensitivity and resolution, and converts individual compounds into corresponding peaks, thus it can increase the probability of discovery of potential novel metabolites.7)
In view of efficiency and simplicity, HPLC was performed as the main analytical technique for the determination of contents of saponins in PJR.5,10) However, there are some limitations exist in HPLC. Due to the complexity and diversity of the chemical structures, some saponins with similar structure or properties cannot be separated well; in addition, minor or trace saponins are difficultly detected by an UV detector or it needs a quite long time to analyze.13) In order to rapidly determine the saponins in PJR, a rapid and sensitive method, which can effectively address the above issues, should be established.13)
To date, LC-MS has become a strongly analyzing method to identify and quantify the complex composition of Chinese medicine, which achieves high separation, good sensitivity and resolution, and high-speed detection. Compared with MS, triple quadrupole-time of flight tandem mass spectrometry (triple TOF-MS/MS) has higher resolution, accuracy and sensitivity for the determination of precursor and product ions, which is beneficial to determination of the exact molecular weight and chemical structure. While triple quadrupole-linear ion trap tandem (QTRAP)-MS/MS has the functions of triple quadrupole (Q) and linear ion trap (TRAP). In the mode of TRAP, its sensitivity is tens or even hundreds of times higher than an ordinary triple quadrupole; In the mode of Q, it still holds scan modes of ordinary triple quadrupole, such as Product ion Scan (PROS), Precursor ion Scan (PRES), Neutral Loss Scan (NLS) and Multiple Reaction Monitoring (MRM) or Select Reaction Monitoring (SRM). In view of the fact that triple TOF-MS/MS has strong ability of qualitative analysis for full scan, of which the molecular weight can be accurate to four decimal places, and QTRAP-MS/MS has the advantage of quantitative sensitivity and stability when MRM was chosen for quantitation, we chose them for the qualitative and quantitative analysis, respectively. In this study, an efficient and sensitive method based on ultra-fast liquid chromatography (UFLC)-Triple TOF-MS/MS was established to identify 53 saponins in PJR. The regular fragmentation pathways of different types of saponins were also preliminarily deduced. Furthermore, an efficient and sensitive method based on ultra-fast liquid chromatography coupled with UFLC-QTRAP-MS/MS was established and validated for the determination of the contents of 13 saponins in PJR, including notoginsenoside R1, R2 (20S), ginsenoside Re, Rg1, Rb1, Rg2 (20S), Rc, Ro, Rd, pseudoginsenoside F11, RT1, chikusetsusaponin IV and IVa. This validated method was also conducted to quantify 13 saponins in PJR samples harvested at different times, and the quality of samples was evaluated by grey relational analysis (GRA).
The reference substances of notoginsenoside R1, ginsenoside Re, Rg1, Rf, Rb1, Rg2 (20S), Rh1 (20S), Rc, Rh1 (20R), Ro, Rb3, Rd, F4, F2, Rg3 (20R), Rk1, Rg5, CK, pseudoginsenoside F11, RT1, chikusetsusaponin IV and IVa were supplied by Liangwei Bio-technology Co., Ltd. (Nanjing, China). Notoginsenoside R2 (20S) were supplied by Yuanye Bio-technology Co., Ltd. (Shanghai, China). The purity of all above reference substances was more than 98% determined by HPLC. Methanol, acetonitrile and formic acid of HPLC grade were obtained from Merck (Darmstadt, Germany). Ultra-pure water was obtained from a Milli-Q Ultra-pure water system (Millipore, Bedford, MA, U.S.A.). Other reagents were of analytical grade.
Plant MaterialsPJR samples (five-years old) were collected from the cultivation base of Enshi city (Hubei province, China, 30°3′49″N, 109°51′41″E) at different harvesting times. These samples (PJR01–PJR05) were harvested on September 10, September 17, October 20, November 1 and November 14 in 2017, separately. The botanical origin of the samples was authenticated by Professor Xunhong Liu (Department for Authentication of Chinese Medicine, Nanjing University of Chinese Medicine, China), and the voucher specimens were deposited at the Herbarium in Nanjing University of Chinese Medicine.
Preparation of Standard SolutionsTwenty three saponins (notoginsenoside R1, R2 (20S), ginsenoside Re, Rg1, Rf, Rb1, Rg2 (20S), Rh1 (20S), Rc, Rh1 (20R), Ro, Rb3, Rd, F4, F2, Rg3 (20R), Rk1, Rg5, CK, pseudoginsenoside F11, RT1, chikusetsusaponin IV and IVa) and 13 saponins (notoginsenoside R1, R2 (20S), ginsenoside Re, Rg1, Rb1, Rg2 (20S), Rc, Ro, Rd, pseudoginsenoside F11, RT1, chikusetsusaponin IV and IVa, as shown in Fig. 1) reference compounds were dissolved in 70% MeOH for UFLC-Triple TOF-MS/MS and UFLC-QTRAP-MS/MS analysis, respectively.
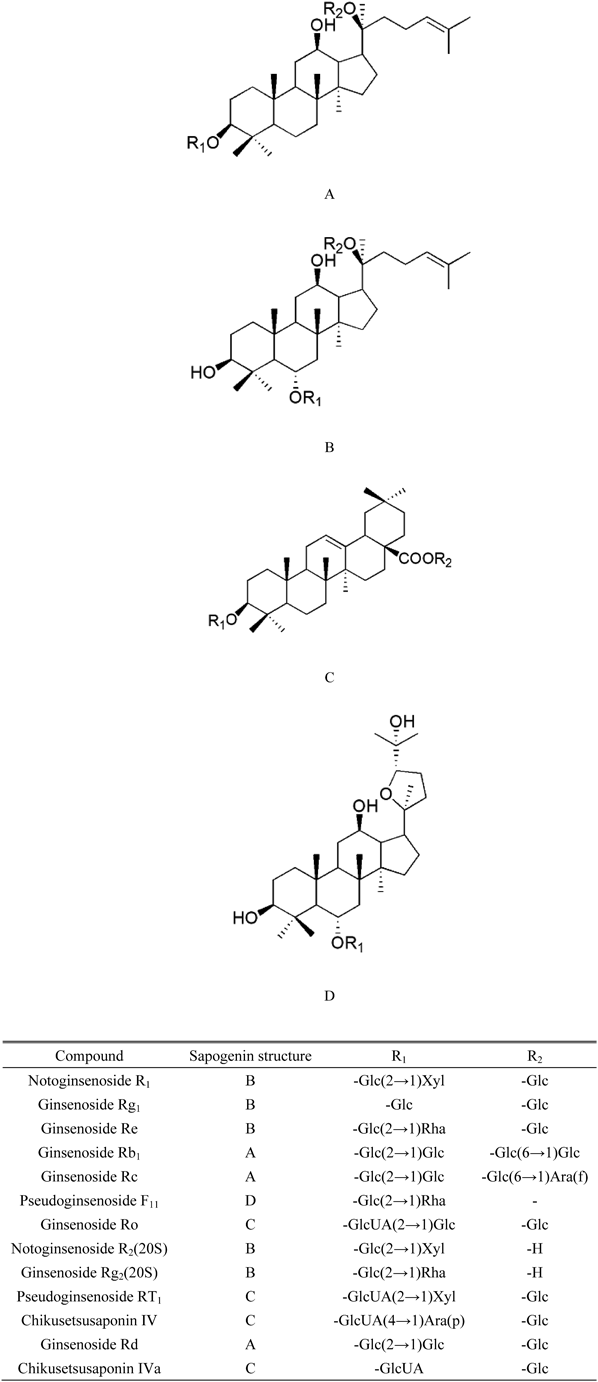
(A) The aglycone of protopanaxadiol (PPD) saponins; (B) the aglycone of protopanaxatriol (PPT) saponins; (C) the aglycone of oleanolic acid (OA) saponins; (D) the aglycone of ocotillone (OC) saponins. GlcUA, β-D-glucuronic acid, Ara (p), α-L-arabinose (pyranose); Ara (f), α-L-arabinose (furanose); Rha, α-L-rhamnose; Xyl, β-D-xylose.
Approximately 0.5 g of PJR powder (60 mesh) was accurately weighed, dissolved in 10 mL of 70% methanol and ultrasonically extracted (40 kHz, 500 W) for 60 min. After cooling to room temperature, the sample solution was replenished to its original weight with 70% methanol. The extraction was centrifuged for 10 min at 12000 r/min (8050 g) and then filter through a microporous membrane (0.22 µm, Jinteng laboratory equipment Co., Ltd., Tianjin, China) prior to UFLC-Triple TOF-MS/MS and UFLC-QTRAP-MS/MS analysis.
UFLC-Triple TOF-MS/MS AnalysisUFLC analysis was performed on an UFLC-20ADXR system (Shimadzu, Kyoto, Japan). Chromatographic separation of 23 saponins in PJR was obtained on a Synergi™ Hydro-RP 100 Å column (2.0 × 100 mm, 2.5 µm, Phenomenex, U.S.A.) with 0.1% formic acid aqueous solution (A)–0.1% formic acid acetonitrile (B). The elution condition was optimized as follows: 0–10 min, 10–35% B; 10–14 min, 35–42% B; 14–24 min, 42–95% B. The temperature was 40°C, the flow rate was 0.4 mL/min, the inject volume was 2.0 µL.
MS data were recorded by a Triple TOF™ 5600 System-MS/MS High Resolution Quadrupole Time-of-Flight Mass Spectrometer (AB Sciex, Framingham, MA, U.S.A.), which was equipped with an electrospray ionization (ESI) source. The negative ion mode was adopted by this study. The mass scanning range was from m/z 50 to 1500. The optimized parameters were obtained as follows: the ion source temperature, 550°C; the flow rate of curtain gas, 40 L/min; the flow rate of nebulization gas, 55 L/min; the flow rate of auxiliary gas, 55 L/min; the spray voltage, −4500 V; the declustering voltage, −100 V. All MS data were acquired by PeakView 1.2 software.
UFLC-QTRAP-MS/MS AnalysisUFLC analysis was performed on an UFLC-20ADXR system (Shimadzu). Chromatographic separation of 13 saponins in PJR was obtained on a XBridge®C18 column (4.6 × 100 mm, 3.5 µm, Waters, Ireland) with 0.1% formic acid aqueous solution (A)–0.1% formic acid acetonitrile (B). The elution condition was optimized as follows: 0–1 min, 25% B; 1–1.5 min, 25–40% B; 1.5–4 min, 40% B; 4–5 min, 40–98% B; 5–5.5 min, 98–2% B; 5.5–8.1 min, 2% B. The temperature was 30°C, the flow rate was 0.8 mL/min, the inject volume was 2.0 µL.
MS data was recorded by a QTRAP 4500 Triple Quadrupole Linear Ion Trap Mass Spectrometer (AB Sciex, Framingham, MA, U.S.A.), which was equipped with an ESI source. MRM mode was chosen for detection of the specific ion. The optimized parameters were obtained as follows: the ion source temperature, 650°C; the flow rate of curtain gas, 30 L/min; the flow rate of nebulization gas, 65 L/min; the flow rate of auxiliary gas, 65 L/min; the spray voltage, 5500 V in the positive mode and −4500 V in the negative mode. All MS data were acquired by Analyst 1.6.3 software.
Validation of QuantificationLinearity, Limit of Detection, and Limit of QuantificationThe calibration curves were constructed by diluting the stock solutions to appropriate concentrations with 70% methanol, and were carried out by plotting the peak area versus the corresponding concentration of 13 saponins. The limits of detection (LOD) and quantification (LOQ) were determined according to signal-to-noise ratio (S/N) of about 3 and 10, separately.
Precision, Repeatability, Stability, and RecoveryThe precision of the developed method was determined by intra- and inter-day variations. The known concentrations of 13 standard saponins solutions were determined for six duplicates in a day and three consecutive days, separately. For the repeatability test, six replicates of the same PJR powder were ultrasonically extracted and determined by UFLC-QTRAP-MS/MS. The contents of 13 saponins in the sample solution were determined at 0, 2, 4, 8, 12 and 24 h for the stability test. For the recovery test, 0.25 g sample powder was added certain amounts of each reference solution. The amount of reference solution was at three levels of 80, 100 and 120% of the known content of the sample, which were repeated three times for each level. The average recovery was calculated as follows: recovery (%) = (m1 − m2)/m3 × 100%. m1 is the measured amount, m2 is the original amount and m3 is the added amount.
Grey Relational AnalysisThe quality of PJR samples was evaluated by GRA, as follows: Supposing that n samples, each of which has m indexes, formed the sequence {Xik} (i = 1, 2, 3, ..., n; k = 1, 2, 3, ..., m; in this study, n = 5, m = 13). According to formula (1), this study normalized the data of PJR samples.
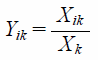 | (1) |
(Yik: normalized data, Xik: original data, Xk: average value of the kth index of nth samples).
While evaluating with GRA, the optimal and worst sequences were set as {Xsk} and {Xtk}, separately. Then, the correlation coefficient of the optimal sequence:
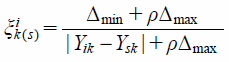 | (2) |
Δmin = min|Yik − Ysk|, Δmax = max|Yik − Ysk|, Ysk: The maximum value of Yik corresponding to each compound, ρ = 0.5.
The correlation coefficient of the worst sequence:
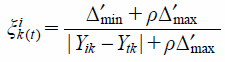 | (3) |
Δ′min = min|Yik − Ytk|, Δ′max = max|Yik − Ytk|, Ytk: The minimum value of Yik corresponding to each compound, ρ = 0.5.
The correlation degree of the optimal sequence:
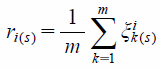 | (4) |
The correlation degree of the worst sequence:
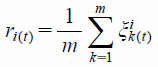 | (5) |
The relative correlation degree:
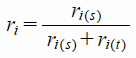 | (6) |
The extraction conditions including extraction solvent (50% methanol–ethanol, 70% methanol–ethanol, 90% methanol–ethanol and water, respectively), ultrasonic time (30, 60 and 90 min, respectively) and ratios of sample to solvent (1 : 10, 1 : 20 and 1 : 30, respectively) were compared for the efficient extraction of saponins in PJR. Seventy percent methanol extract showed more chromatographic peaks and higher content of Chikusetsusaponin IVa; the extract of 1 : 20 and 60 min showed more chromatographic peaks and higher content of Chikusetsusaponin IVa. It was found that the optimum extraction conditions were 70% methanol at a ratio of 1 : 20 for 60 min.
Optimization of UFLC-Triple TOF-MS/MS and UFLC-QTRAP-MS/MS ConditionsDifferent mobile phases (aqueous solution–methanol–acetonitrile, 0.1% formic acid aqueous solution–acetonitrile and 0.1% formic acid aqueous solution–0.1% formic acid acetonitrile, respectively) were compared for the good separation and peak pattern of saponins. The result demonstrated that 0.1% formic acid aqueous solution–0.1% formic acid acetonitrile was the more suitable mobile phase. Different detection modes were optimized for UFLC-Triple TOF-MS/MS method. The result showed that more information on active ingredients was obtained in the negative ion mode than in the positive ion mode. The optimized MS/MS parameters of UFLC-QTRAP-MS/MS method were shown in Table 1, and MRM of 13 saponins was presented in Fig. 2.
| Compound | tR (min) | MRM parameters | |||
|---|---|---|---|---|---|
| MRM transitions (m/z) | DP (V) | CE (eV) | Ionization mode | ||
| Notoginsenoside R1 | 2.01 | 932.412/638.300 | −225.00 | −50.00 | ESI− |
| Ginsenoside Rg1 | 2.66 | 845.680/637.400 | −20.00 | −35.00 | ESI− |
| Ginsenoside Re | 2.68 | 991.700/799.400 | −120.00 | −45.00 | ESI− |
| Ginsenoside Rb1 | 3.50 | 1109.600/352.00 | 111.00 | 31.00 | ESI+ |
| Ginsenoside Rc | 3.70 | 1077.600/1077.600 | −120.00 | −60.00 | ESI− |
| Pseudoginsenoside F11 | 3.90 | 845.400/653.400 | −110.00 | −50.00 | ESI− |
| Ginsenoside Ro | 4.00 | 955.585/793.400 | −5.00 | −60.00 | ESI− |
| Notoginsenoside R2(20S) | 4.10 | 769.427/637.400 | −25.00 | −38.00 | ESI− |
| Ginsenoside Rg2(20S) | 4.30 | 829.600/637.400 | −20.00 | −40.00 | ESI− |
| Pseudoginsenoside RT1 | 4.40 | 949.380/641.300 | 261.00 | 55.00 | ESI+ |
| Chikusetsusaponin IV | 4.40 | 926.000/569.300 | −15.00 | −60.00 | ESI− |
| Ginsenoside Rd | 4.50 | 991.700/783.400 | −120.00 | −55.00 | ESI− |
| Chikusetsusaponin IVa | 4.90 | 793.530/631.200 | −185.00 | −58.00 | ESI− |

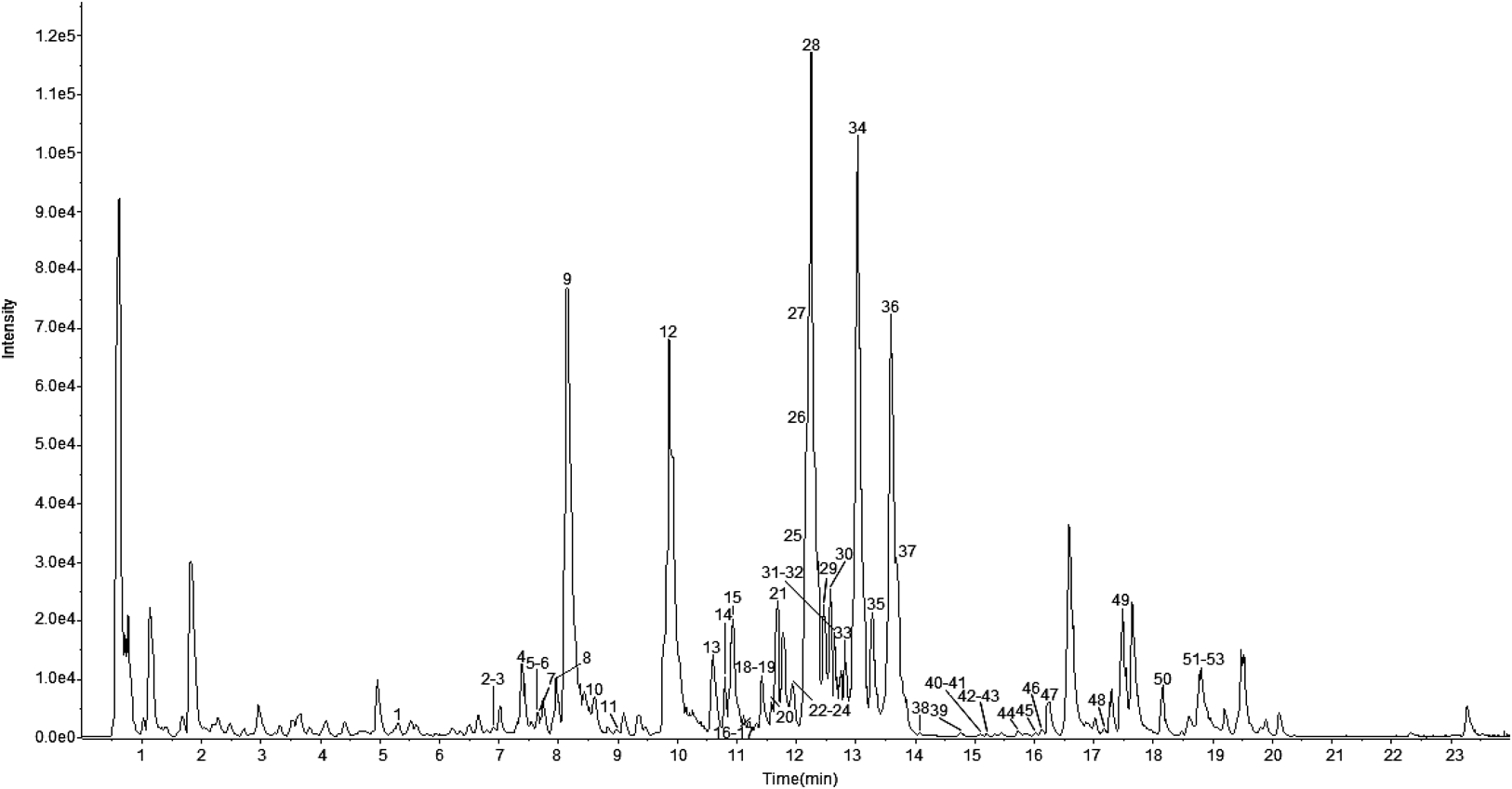
Twenty three reference compounds and 70% methanol extract of PJR were analyzed by UFLC-Triple TOF-MS/MS under the optimized conditions. The base peak chromatogram (BPC) of 70% methanol extract of Panacis Japonici Rhizoma (PJR03) in the negative ion mode was shown in Fig. 3. According to the retention time, precursor ions and product ions, compared with the reference compounds and related literatures, 53 saponins were identified, including 23 saponins unambiguously identified by reference compounds. Identification of 53 saponins in Panacis Japonici Rhizoma by UFLC-Triple TOF-MS/MS was shown in Table 2. In addition, the regular fragmentation pathways were preliminarily deduced by fragmentation behavior of 53 saponins analyzed in the negative mode.
| No. | tR (min) | Formula | Measured mass (m/z) | Error (10−6) | MS/MS fragmentation (m/z) | Compound | References |
|---|---|---|---|---|---|---|---|
| 1 | 5.28 | C48H82O20 | 1023.5414[M + HCOO]− | 4.2 | 977.5414 [M−H]−, 653.4309 [M−H−2Glc]− | Yesanchinoside B | 14 |
| 2 | 6.91 | C42H72O15 | 861.4865 [M+HCOO]− | 2.7 | 815.4833 [M−H]−, 653.4295 [M−H−Glc]−, 491.3744 [M−H−2Glc]− | Majonoside R1(Majoroside R1) | 15–18 |
| 3 | 6.93 | C54H92O23 | 1153.6045 [M+HCOO]− | 3.8 | 1107.6025 [M−H]− | Ginsenoside B1 | 19 |
| 4 | 7.34 | C48H82O19 | 961.5393 [M−H]− | 1.7 | 799.4891 [M−H−Glc]−, 637.4345 [M−H−2Glc]− | Notoginsenoside N/M/R6/R3/20-Glc-ginsenoside-Rf/Ginsenoside Re1/Re2/Re3 | 20–25 |
| 5 | 7.6 | C53H90O22 | 1077.5860 [M−H]− | 0.8 | 945.5442 [M−H−Xyl]− | Vinaginsenoside R7 | 17, 21 |
| 6 | 7.64 | C47H80O18 | 977.5351 [M+HCOO]− | 3.7 | 931.5333 [M−H]−, 799.4879 [M−H−Xyl]−, 637.4334 [M−H−Xyl−Glc]− | Notoginsenoside R1a) | 20, 22, 23, 25, 26 |
| 7 | 7.67 | C47H80O18 | 977.5340 [M+HCOO]− | 2.6 | 931.5334 [M−H]−, 799.4879 [M−H−Xyl]−, 637.4342 [M−H−Xyl−Glc]- | Ginsenoside Re4/Quinquenoside L17 | 17, 24, 25 |
| 8 | 8.05 | C48H82O18 | 991.5519 [M+HCOO]− | 4.7 | 945.5492 [M−H]−, 799.4876 [M−H−Rha]−, 783.4932 [M−H−Glc]−, 637.4314 [M−H−Rha−Glc]− | Ginsenoside Rea) | 16, 23, 26–28 |
| 9 | 8.33 | C42H72O14 | 845.4930 [M+HCOO]− | 4.4 | 799.4890 [M−H]−, 637.4346 [M−H−Glc]−, 475.3792 [M−H−2Glc]− | Ginsenoside Rg1a) | 16, 23, 25, 28 |
| 10 | 8.71 | C45H74O17 | 885.4884 [M−H]− | 3.5 | 841.5011 [M−H−CO2]−, 799.4914 [M−H−Ma]−, 781.4791 [M−H−Ma−H2O]−, 637.4352 [M−H−Ma−Glc]−, 619.4225 [M−H−Ma−H2O−Glc]−, 475.3788 [M−H−Ma−2Glc]− | Malonyl-ginsenoside Rg1 | 16, 20, 22, 26, 29 |
| 11 | 9.05 | C55H92O23 | 1119.5977 [M−H]− | 1.8 | 1077.5946 [M−H−Ac]−, 1059.5849 [M−H−Ac−H2O]− | Ginsenoside Rs1/Rs2 | 23, 25, 26, 30 |
| 12 | 10.07 | C49H78O19 | 1015.5142 [M+HCOO]− | 3.3 | 969.4790 [M−H]−, 807.4215 [M−H−Glc]− | Chikusetsusaponin V Methyl ester | 31, 32 |
| 13 | 10.61 | C41H70O14 | 831.4777 [M+HCOO]− | 4.8 | 785.4732 [M−H]−, 653.4299M−H−Xyl]− | Majonoside R2 (Majoroside R2)/Pseudoginsenoside RT2 | 28, 33 |
| 14 | 10.87 | C42H72O14 | 845.4931 [M+HCOO]− | 4.3 | 799.4900 [M−H]−, 653.4266 [M−H−Rha]− | Pseudoginsenoside F11a) | 17, 27, 28 |
| 15 | 10.91 | C42H72O14 | 845.4922 [M+HCOO]− | 3.4 | 799.4880 [M−H]−, 637.4340 [M−H−Glc]−, 475.3799 [M−H−2Glc]− | Ginsenoside Rf1) | 22, 23, 26, 27 |
| 16 | 11.33 | C44H74O15 | 887.5029 [M+HCOO]− | 3.4 | 841.4998 [M−H]−, 799.4889 [M−H−Ac]−, 781.4744 [M−H−Ac−H2O]−, 679.45351 [M−H−Glc]−, 637.4275 [M−H−Ac−Glc]−, 619.4137 [M−H−Ac−Glc−H2O]−, 475.3809 [M−H−Ac−2Glc]− | Acetyl-ginsenoside Rg1 | 15, 23 |
| 17 | 11.36 | C41H70O13 | 815.4809 [M+HCOO]− | 2.6 | 769.4792 [M−H]−, 637.4343 [M−H−Xyl]−, 475.3787 [M−H−Xyl−Glc]− | Notoginsenoside R2(20S) a) | 21, 25–28 |
| 18 | 11.42 | C41H70O13 | 769.4750 [M−H]− | 0.8 | 637.4328 [M−H−Xyl/Ara(p)]−, 475.3803 [M−H−Xyl/Ara(p)−Glc]− | Pseudoginsenoside RT3/Ginsenoside F3 | 16, 23, 26 |
| 19 | 11.45 | C53H88O23 | 1137.5663 [M+HCOO]− | −2.2 | 1091.6049 [M−H]−, 929.5554 [M−H−Glc]− | Yesanchinoside G | 21, 34 |
| 20 | 11.5 | C41H70O13 | 815.4812 [M+HCOO]− | 2.9 | 769.4793 [M−H]−, 637.4339 [M−H−Ara]−, 475.3801 [M−H−Ara−Glc]− | Sanchirhinoside A3/A4(20S) | 18, 35 |
| 21 | 11.68 | C54H92O23 | 1107.5971 [M−H]− | 1.3 | — | Ginsenoside Rb11) | 23, 26, 28, 30 |
| 22 | 11.89 | C56H94O24 | 1149.6067 [M−H]− | 0.4 | 1107.6044 [M−H−Ac]−, 1089.5962 [M−H−Ac−H2O]− | Quinquenoside R1 | 15, 23, 25 |
| 23 | 11.9 | C56H94O24 | 1195.6111 [M+HCOO]− | 0.4 | 1149.6158 [M−H]−, 1107.6068 [M−H−Ac]−, 1089.5972 [M−H−Ac−H2O]− | Yesanchinoside F | 14 |
| 24 | 11.93 | C42H72O13 | 783.4881 [M−H]− | −2.4 | 637.4355 [M−H−Rha]−, 475.3802 [M−H−Rha−Glc]− | Ginsenoside Rg2(20S) a) | 16, 26–28, 30, 36 |
| 25 | 12.09 | C36H62O9 | 683.4387 [M+HCOO]− | 3.2 | 637.4411 [M−H]−, 475.3848 [M−H−Glc]− | Ginsenoside Rh1(20S) a) | 16, 26, 27, 36 |
| 26 | 12.12 | C53H90O22 | 1077.5936 [M−H]− | 7.9 | — | Ginsenoside Rca) | 23, 25, 26, 28, 30 |
| 27 | 12.21 | C48H76O19 | 955.4934 [M−H]− | 2.7 | 793.4412 [M−H−Glc]− | Tuberoside A/Hemsgiganoside B | 15, 37, 38 |
| 28 | 12.26 | C36H62O9 | 683.4397 [M+HCOO]− | 4.7 | 637.4350 [M−H]−, 475.3942 [M−H−Glc]− | Ginsenoside Rh1(20R) a) | 29, 30, 33, 39 |
| 29 | 12.29 | C48H76O19 | 1001.4995 [M+HCOO]− | 4.3 | 955.4962 [M−H]− | Ginsenoside Roa) | 26, 28, 30 |
| 30 | 12.61 | C53H90O22 | 1077.5926 [M−H]− | 7.0 | 945.5437 [M−H−Xyl]−, 783.4992 [M−H−Xyl−Glc]−, 765.4824 [M−H−Xyl−Glc−H2O]−, 621.4481 [M−H−Xyl−2Glc]− | Ginsenoside Rb3a) | 23, 27, 28, 30 |
| 31 | 12.62 | C47H74O18 | 925.4812 [M−H]− | 1.1 | 763.4243 [M−H−Glc]−, 569.3858 [M−H−Glc−Ara(f)−H2O−CO2]− | Stipuleanoside R1/Chikusetsusaponin Ib | 15, 40 |
| 32 | 12.82 | C44H74O15 | 887.5011 [M+HCOO]− | 1.4 | 841.4992 [M−H]−, 799.4891 [M−H−Ac]−, 635.4206 [M−H−Ac−Glc]− | Yesanchinoside D | 14 |
| 33 | 12.85 | C47H74O18 | 925.4819 [M−H]− | 1.8 | — | Pseudoginsenoside RT1a) | 6, 40 |
| 34 | 13.06 | C47H74O18 | 925.4835 [M−H]− | 3.6 | 613.3758 [M−H−Glc−Ara−H2O]−, 569.3853 [M−H−Glc−Ara−H2O−CO2]− | Chikusetsusaponin IVa) | 41 |
| 35 | 13.38 | C48H82O18 | 991.5502 [M+HCOO]− | 3.0 | 945.5507 [M−H]− | Ginsenoside Rda) | 16, 23, 28, 30 |
| 36 | 13.55 | C42H66O14 | 793.4399 [M−H]− | 2.4 | 631.3883 [M−H−Glc]− | Chikusetsusaponin IVaa) | 16, 22 |
| 37 | 13.66 | C42H66O14 | 793.4397 [M−H]− | 2.1 | 631.3883 [M−H−Glc]−, 569.3871 [M−H−Glc−CO2−H2O]− | Zingibroside R1/Cynarasaponin C | 16, 33, 37 |
| 38 | 14.05 | C48H82O18 | 991.5489 [M+HCOO]− | 1.7 | 945.5524 [M−H]−, 783.4965 [M−H−Glc]− | Notoginsenoside K | 20 |
| 39 | 14.72 | C36H62O9 | 683.4371 [M+HCOO]− | 0.9 | 637.4319 [M−H]−, 475.3819 [M−H−Glc]− | Ginsenoside F1 | 16, 26 |
| 40 | 15.02 | C42H70O13 | 827.4814 [M+HCOO]− | 3.1 | 781.4800 [M−H]−, 619.4273 [M−H−Glc]− | Ginsenoside Rg9(20E/Z) | 26 |
| 41 | 15.07 | C47H80O17 | 961.5365 [M+HCOO]− | −0.2 | 915.5379 [M−H]−, 783.4967 [M−H−Xyl]−, 621.4476 [M−H−Xyl−Glc]− | Chikusetsusaponin III/Vinaginsenoside R16/R17 | 13, 15, 41 |
| 42 | 15.09 | C47H80O17 | 961.5397 [M+HCOO]− | 3.1 | 915.5964 [M−H]−, 783.4861 [M−H−Ara(f)]−, 621.4389 [M−H−Ara(f)−Glc]− | Notoginsenoside Fe | 15–17, 22 |
| 43 | 15.13 | C48H82O17 | 975.5547 [M+HCOO]− | 2.5 | 929.5554 [M−H]− | Vinaginsenoside R3 | 16 |
| 44 | 15.78 | C42H70O12 | 811.4866 [M+HCOO]− | 3.5 | 765.4858 [M−H]−, 619.4269 [M−H−Rha]− | Ginsenoside Rg6 | 15, 16, 30 |
| 45 | 16.07 | C42H70O12 | 811.4882 [M+HCOO]− | 5.4 | 765.4839 [M−H]−, 619.4253 [M−H−Rha]− | Ginsenoside F4a) | 16, 22 |
| 46 | 16.36 | C42H72O13 | 829.4977 [M+HCOO]− | 4.0 | 783.4965 [M−H−Glc]−, 621.4385 [M−H−Glc]−, 459.3825 [M−H−2Glc]− | Ginsenoside F2a) | 17, 21, 26, 30 |
| 47 | 17.22 | C44H74O14 | 871.5091 [M+HCOO]− | 4.7 | 825.5070 [M−H]−, 783.4944 [M−H−Ac]−, 765.4914 [M−H−Ac−H2O]−, 621.4480 [M−H−Ac−Glc]− | Ginsenoside Rs3 | 16, 17, 26 |
| 48 | 17.28 | C42H72O13 | 829.4956 [M+HCOO]− | 1.4 | 783.4930 [M−H]−, 621.4397 [M−H−Glc]−, 459.3854 [M−H−2Glc]− | Ginsenoside Rg3(20R) a) | 16, 17, 30, 36 |
| 49 | 17.65 | C41H64O13 | 763.4305 [M−H]− | 4.1 | 613.3769 [M−H−Xyl−H2O]−, 569.3859 [M−H−Xyl−CO2]− | Pseudoginsenoside Rp1 | 6, 42 |
| 50 | 18.37 | C42H70O12 | 811.4855 [M−H]− | 2.1 | 765.4847 [M−H]−, 603.4265 [M−H−Glc]− | Ginsenoside Rk1a) | 21, 26, 27, 30, 36 |
| 51 | 18.85 | C36H58O8 | 663.4117 [M+HCOO]− | 2.1 | 617.4084 [M−H]−, 455.3540 [M−H−Glc]− | Oleanolic acid 28-O-β-D-glucopyranose (PJS-1) | 6, 36 |
| 52 | 18.95 | C42H70O12 | 811.4888 [M+HCOO]− | 6.2 | 765.4855 [M−H]−, 603.4227 [M−H−Glc]− | Ginsenoside Rg5a) | 26, 30, 36 |
| 53 | 19.04 | C36H62O8 | 667.4419 [M+HCOO]− | 0.4 | 621.4397 [M−H]−, 459.3920 [M−H−Glc]− | Ginsenoside CKa) | 21, 22, 26 |
Ac, acetyl (CH3CO); Ma, malonyl (HCOOCH2CO); Glc, β-D-glucose; Ara(p), α-L-arabinose (pyranose); Ara(f), α-L-arabinose (furanose); Rha, α-L-rhamnose; Xyl, β-D-xylose. a) Compounds were identified by comparison with reference compounds.
In this study, [M−H]− and [M+HCOO]− were observed as the precursor ions of saponins. Among them, [M+HCOO]− formed [M−H]− by the loss of HCOOH (46 Da). According to sapogenin structure, these saponins can be divided into four types, which were protopanaxadiol (PPD), protopanaxatriol (PPT), oleanolic acid (OA) and ocotillone (OC) and mostly exhibited fragmentation ways corresponding to losses of H2O, CO2, sugars and other substituents until the formation of characteristic product ions. PPD type of saponins (3, 5, 11, 19, 21, 22, 26, 30, 35, 38, 41, 42, 43, 46, 47, 48, 50 and 52 in Table 2) possessed a characteristic product ion at 459, for example, ginsenoside Rg3 (20R) (Fig. 4A), the fragment ion 459.3814 of which can be considered as the losses of 324 Da (Glc+Glc) (Glc: β-D-glucose). PPT type of saponins (4, 6, 7, 8, 9, 10, 15, 16, 17, 18, 20, 23, 24, 25, 28, 32, 39, 40, 44, 45 and 53 in Table 2) possessed a characteristic product ion at 475, for example, notoginseng R2 (20S) (Fig. 4B), the fragment ion 475.3787 of which can be corresponding to [M−H−Xyl−Glc]− (Xyl: β-D-xylose). OA type of saponins (12, 27, 29, 31, 33, 34, 36, 37, 49 and 51 in Table 2) possessed a characteristic product ion at 569, for example, Chikusetsusaponin IV (Fig. 4C), the fragment ion 569.3853 of which could be considered the losses of 356 Da (Glc+Ara+H2O+CO2) (Ara: α-L-arabinose). OC type of saponins (1, 2, 13 and 14 in Table 2) possessed a characteristic product ion at 653, for example, pseudoginsenoside F11 (Fig. 4D), the fragment ion 653.4266 of which corresponded to [M−H−Rha]− (Rha: α-L-rhamnose). And sometimes it might form a product ion of 491 by a loss of a glucose unit. Based on these comparative analysis of fragments, these characteristic sugar fragments were mostly observed by simultaneous or successive losses of 162 Da (-Glc), and/or 132 Da (-Ara or -Xyl), and/or 146 Da (-Rha). However, the amount and the type of sugar units were observed different in the fragmentation process. The lost masses of 179, 161, 119, 113 and 101, might correspond to a Glc (fragment ions 179, 161, 119, 113 and 101 observed from the MS/MS spectra of ginsenoside Rb1, CK and F2, etc.), while 149 and 131 were possibly corresponding to Ara or Xyl (fragment ions 149 and 131 observed from the MS/MS spectra of ginsenoside Rc and Rb3). The product ions of 323 and 221, was possibly consistent with Glc-Glc (fragment ions 323 and 221 observed from the MS/MS spectra of ginsenoside Rb1), while 191 was possibly consistent with Glc-Ara or Glc-Xyl (fragment ion 191 observed from the MS/MS spectra of ginsenoside Rc and Rb3).13,43,44)
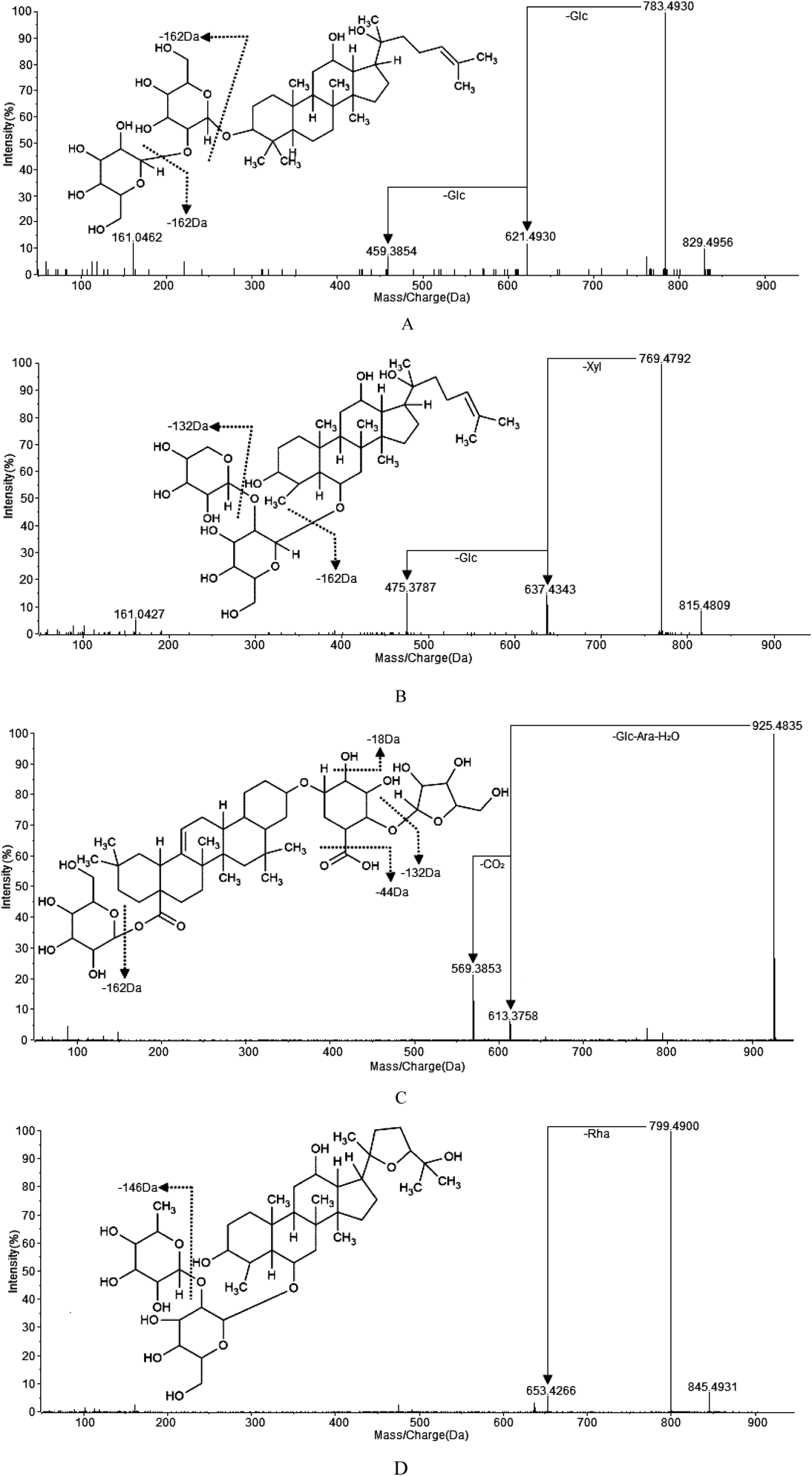
(A) Ginsenoside Rg3(20S), which has the aglycone of protopanaxadiol (PPD) saponins; (B) Notoginseng R2(20S), which has the aglycone of protopanaxatriol (PPT) saponins; (C) Chikusetsusaponin IV, which has the aglycone of oleanolic acid (OA) saponins; (D) Pseudoginsenoside F11, which has the aglycone of ocotillone (OC) saponins.
As shown in Table 3, the calibration curves of saponins with r (correlation coefficient) > 0.999, it showed that linear relationships of each compound were good within the linear range. The range of LOD and LOQ were 0.25–6.65 ng/mL and 0.75–20.33 ng/mL, respectively. As shown in Table 4, the RSDs of intra- and inter-day variations of 13 compounds were all less than 3.2%. For the repeatability test, RSDs ranged from 1.5 to 3.5%, which indicated that the method had good repeatability. The stability results of saponins were expressed as RSDs from 1.1 to 5.0%, which demonstrated that the sample solution was stable within 24 h. The average recovery varied from 98.77 to 101.7%, and the range of RSDs was 1.0–3.1%.
| Compound | Linear equationsa) | r | Linear range (ng/mL) | LOD (ng/mL) | LOQ (ng/mL) |
|---|---|---|---|---|---|
| Notoginsenoside R1 | y = 8.96x + 222 | 0.9996 | 12.7–10140 | 2.54 | 7.62 |
| Ginsenoside Rg1 | y = 54.2x + 11500 | 0.9993 | 7.23–231200 | 1.45 | 4.35 |
| Ginsenoside Re | y = 3.85x + 3170 | 0.9994 | 33.3–532000 | 6.65 | 19.95 |
| Ginsenoside Rb1 | y = 54.3x + 3950 | 0.9999 | 12.5–100200 | 2.51 | 7.53 |
| Ginsenoside Rc | y = 27.5x − 280 | 0.9997 | 12.5–4990 | 0.25 | 0.75 |
| Pseudoginsenoside F11 | y = 140x + 18500 | 0.9994 | 19.2–123200 | 3.84 | 13.25 |
| Ginsenoside Ro | y = 15.4x + 23400 | 0.9991 | 61.2–244000 | 4.90 | 16.20 |
| Notoginsenoside R2(20S) | y = 3.47x + 1100 | 0.9995 | 6.78–27100 | 1.36 | 4.08 |
| Ginsenoside Rg2(20S) | y = 202x + 4260 | 0.9991 | 3.35–10720 | 0.67 | 2.01 |
| Pseudoginsenoside RT1 | y = 7.97x + 12900 | 0.9991 | 31.2–1000000 | 6.24 | 18.72 |
| Chikusetsusaponin IV | y = 16.9x + 1170 | 1.0000 | 110–440000 | 6.13 | 20.33 |
| Ginsenoside Rd | y = 15.6x + 10400 | 0.9999 | 31.4–126000 | 6.28 | 18.84 |
| Chikusetsusaponin IVa | y = 64.2x + 89100 | 0.9993 | 8.29–332000 | 0.81 | 2.43 |
LOD, limit of detection; LOQ, limit of quantification. a) y, peak area; x, concentration (ng/mL).
| Compound | Precision (RSD, %) | Repeatability (RSD, %) (n = 6) | Stability (RSD, %) (n = 6) | Recovery | ||
|---|---|---|---|---|---|---|
| Intra-day (n = 6) | Inter-day (n = 3) | Average (%) | RSD (%) | |||
| Notoginsenoside R1 | 2.0 | 2.1 | 1.5 | 1.5 | 99.76 | 3.1 |
| Ginsenoside Rg1 | 2.1 | 2.4 | 2.0 | 1.6 | 98.84 | 1.7 |
| Ginsenoside Re | 2.1 | 2.7 | 2.4 | 1.8 | 99.77 | 1.0 |
| Ginsenoside Rb1 | 3.0 | 2.6 | 2.9 | 1.5 | 100.8 | 2.5 |
| Ginsenoside Rc | 3.2 | 2.9 | 1.8 | 1.7 | 99.32 | 2.7 |
| Pseudoginsenoside F11 | 3.0 | 3.2 | 2.2 | 3.1 | 98.77 | 2.2 |
| Ginsenoside Ro | 2.9 | 2.5 | 2.8 | 2.0 | 99.63 | 2.4 |
| Notoginsenoside R2 (20S) | 3.0 | 2.8 | 3.5 | 5.0 | 99.09 | 2.6 |
| Ginsenoside Rg2 (20S) | 3.1 | 2.6 | 2.8 | 2.7 | 98.93 | 1.5 |
| Pseudoginsenoside RT1 | 2.2 | 2.4 | 1.8 | 1.1 | 101.7 | 2.7 |
| Chikusetsusaponin IV | 3.0 | 2.4 | 2.2 | 2.9 | 100.2 | 2.8 |
| Ginsenoside Rd | 2.8 | 2.8 | 3.2 | 3.2 | 99.12 | 2.9 |
| Chikusetsusaponin IVa | 2.7 | 2.7 | 2.2 | 2.5 | 100.1 | 2.1 |
RSD, relative standard deviation.
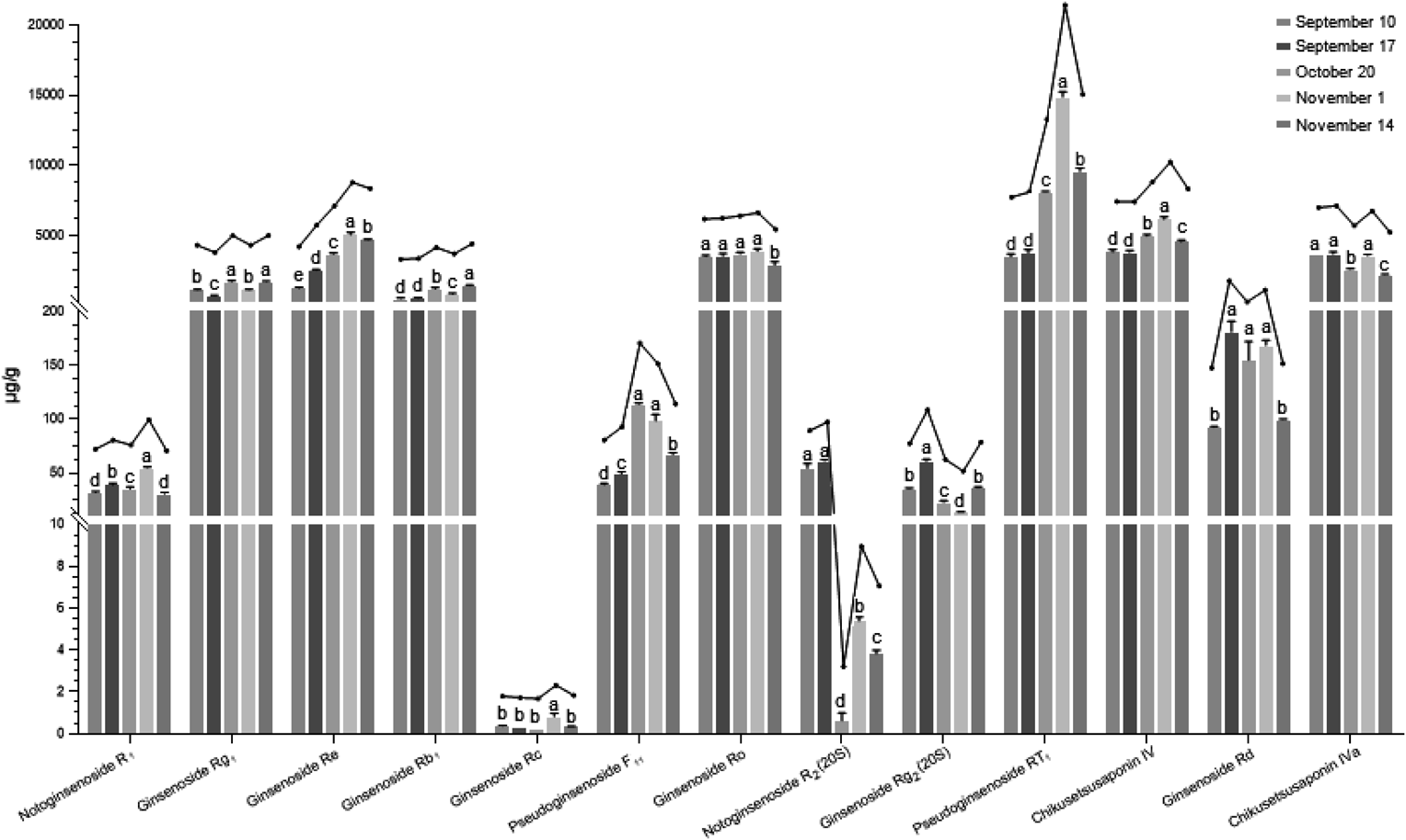
Thirteen saponins (contain four types of saponins) was chosen as the quantitative standards based on extensive existence of relatively high concentration in PJR or biological potential significance according to the qualitative results and the related studies.5,45,46) Contents of 13 saponins in PJR samples harvested at different times were determined by the optimized UFLC-QTRAP-MS/MS method. As shown in Fig. 5, the content of each saponin was analyzed by one-way ANOVA. It showed that contents of saponins among different samples demonstrated significant differences. The content of notoginsenoside R2(20S) varied nearly 88-fold, and was more obviously than other saponins, especially between September 17 and October 20. Among these saponins, the content of ginsenoside Re, Ro, pseudoginsenoside F11, RT1 and chikusetsusaponin IV appeared similar trends with harvest time, which went up to reach the maximum, then gradually decreased and the maximum mostly appeared between October 20 and November 1. As shown in Fig. 6, the ginsenoside Ro/Re ratios were found to be different when compared across samples from different harvest time. The average ratios were calculated as 2.79, 1.40, 1.00, 0.77 and 0.62 for PJR samples harvested at different times, respectively. The content of ginsenoside Ro was significantly higher than that of Re on September 10 and September 17, while they were both approximately equal on October 20. However, the content of Ro was significantly lower than the content of Re on November 1 and November 14. Therefore, the ratios of ginsenoside Ro/Re might change along with the harvesting time.
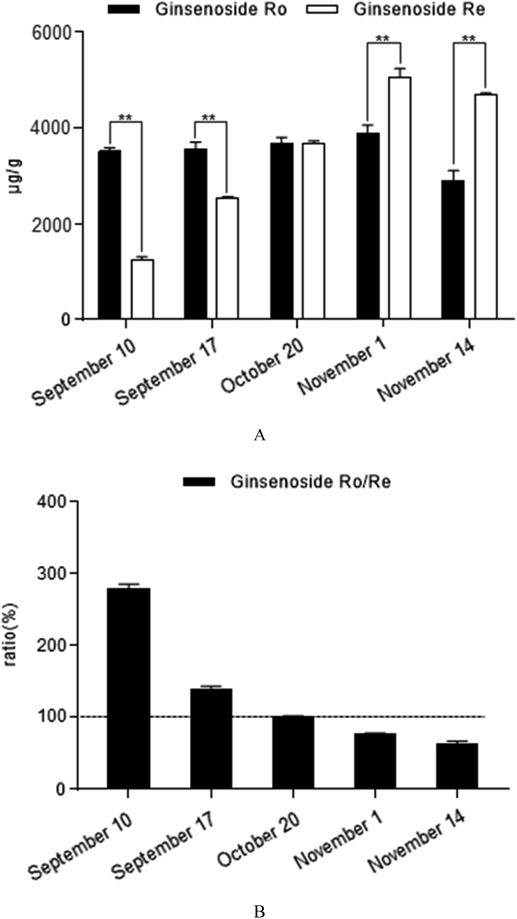
** p < 0.01, independent sample t-test; (B) Ratio of ginsenoside Ro/Re in Panacis Japonici Rhizoma samples harvested at different times.
GRA was applied to evaluating the quality of PJR based on contents of 13 saponins. “Relative correlation degree” was used to describe the correlation relationship between the content of each component and the harvest time. The higher the relative correction degree, the more obvious the relationship, which showed that the overall quality of PJR was relatively better. The result of GRA (Table 5) showed that the comprehensive quality of PJR around October 20 might be better than other time, which was in accordance with the traditional harvested period.
| Items | PJR01 | PJR02 | PJR03 | PJR04 | PJR05 |
|---|---|---|---|---|---|
| Relative correlation degree | 0.5412 | 0.4660 | 0.5973 | 0.5144 | 0.5686 |
| Quality ranking | 3 | 5 | 1 | 4 | 2 |
In conclusion, an efficient and sensitive method based on UFLC-Triple TOF-MS/MS and UFLC-QTRAP-MS/MS was established for qualitative and quantitative analysis of saponins in PJR, respectively. By using UFLC-Triple TOF-MS/MS method, 53 saponins were identified in PJR and 23 of them were unequivocally identified by reference substances. In accordance with the fragmentation behavior of 53 saponins, four types of saponins mostly exhibited fragmentation ways corresponding to losses of H2O, CO2, sugars and other substituents until the formation of sapogenin fragment ions. By UFLC-QTRAP-MS/MS method, 13 saponins were simultaneously determined in PJR samples harvested at different times, and among them, certain saponins content demonstrated some trends. In general, the result of GRA showed the comprehensive quality of PJR harvested around October 20 might be better. This study was meaningful to the quality assessment and control of PJR. What is more, it might provide the basic information for confirming its optimal harvested period.
This research was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions of China (No. ysxk-2014).
The authors declare no conflict of interest.