2019 Volume 67 Issue 8 Pages 775-777
2019 Volume 67 Issue 8 Pages 775-777
Nocardia is a potent bacterial producer of bioactive compounds. From a culture of Nocardia beijingensis NBRC 16342, we isolated four aromatic compounds, named beijinchromes A–D (1–4). We purified them by silica gel chromatography and reverse phase HPLC, and identified their structures by NMR and high resolution (HR)-MS analyses. 1, 2, and 4 are novel 1,2,3,8-tetrasubstituted naphthalenes, and 3 is a novel 3,8-disubstituted ortho-naphthoquinone. 1 and 2 exert antioxidant activities, and 3 exhibits antibiotic activity. Remarkably, the putative biosynthetic gene clusters for 1–4 are widely distributed in 37 Nocardia species, implying their potential to produce this family of compounds and important biological functions of beijinchromes.
Nocardia is one of the actinomycete species that belongs to the same genetic family (Corynebacteriaceae) as the genus Mycobacterium and Corynebacterium, but is apart from the well-known antibiotic producer, Streptomyces.1) They are isolated from a broad range of sources in the environment, including soil, plant, and the human body. Some pathogenetic Nocardia strains cause disorders of the lung, the central nervous system, the brain, and the skin.2) Other strains produce pharmaceutically important antibiotics, such as the immunosuppressive terpenoid brasilicardin A,3) and the macrolide antibiotic brasilinolide A.4) Recent genome sequencing projects suggested that Nocardia is a promising source of secondary metabolites.5) However, their secondary metabolism has not been exploited extensively, considering the fact that the numbers of isolated compounds are much lower than those of secondary metabolite genes.
To seek novel pharmaceutically important bioactive compounds, we screened the metabolites from Nocardia beijingensis6) NBRC 16342, isolated from mud taken from a sewage ditch. Some of the N. beijingensis species are opportunistic bacteria and responsible for human infection.7) Thus, the secondary metabolites from this strain are intriguing from the point of view of both drug discovery and chemical ecology. The antiSMASH8) analysis of the genome of N. beijingensis NBRC 16342 revealed that there are 38 secondary metabolite gene clusters, but there is no report on any secondary metabolite from this species. Therefore, it is likely that this strain possesses a cryptic secondary metabolism that awaits to be discovered. Herein, we report the isolation and structural elucidation of four aromatic metabolites produced by N. beijingensis, named as beijinchromes A–D (1–4, Fig. 1).

N. beijingensis NBRC 16342 was cultured in A-3M medium9) for 6 d, and the resulting culture broth was directly extracted with n-butanol. The crude n-butanol extract was partitioned by repeated flash silica-gel column chromatographies, and 1–4 were finally purified by reverse-phase HPLC (1: 1.4 mg, 2: 2.1 mg, 3: 1.8 mg, 4: 0.7 mg).
The molecular formula of 1 was established as C14H14O4, which was consistent with the 13C-NMR spectrum displaying 14 resonances (Table 1). One dimensional NMR (1D-NMR) and Heteronuclear Multiple Quantum Correlation (HMQC) spectra further indicated that these carbon signals comprise one ketone, six non-protonated aromatic carbons, four aromatic methines, two methyls, and one aliphatic methylene. In addition, 1H-NMR displayed two exchangeable protons (δH 5.78 and 9.12) likely derived from phenolic hydroxyl groups. The position of the methoxy group in ring A was fully established, based on the Correlation Spectroscopy (COSY) and Heteronuclear Multiple Bond Coherence (HMBC) correlations derived from the H5, H6, H7, and methoxy protons, as well as the presence of the trisubstituted benzene ring (Fig. 2). The connection of C4/C4a was supported by the HMBC correlations from H4 and H5, and the connections among C9/C10/C11 were established by the HMBC correlations from H11 (Fig. 2). Moreover, except for the connection of C1/C2 (Fig. 2), the substitution pattern of the second aromatic ring B was established based on the HMBC correlations derived from H4, H9, and the phenolic protons. Finally, the connection of C1/C2 was inferred from the molecular formula of 1, along with the upfield shifted 13C-NMR resonances of C1 and C2 (δC 138.9 and 138.2). Overall, the chemical structure of 1 was determined, as depicted in Fig. 1.
| Beijinchrome A (1) | Beijinchrome B (2) | Beijinchrome C (3) | Beijinchrome D (4) | |||||
|---|---|---|---|---|---|---|---|---|
| Position | δH, mult (J in Hz) | δC, type | δH, mult (J in Hz) | δC, type | δH, mult (J in Hz) | δC, type | δH, mult (J in Hz) | δC, type |
| 1 | — | 138.9, C | — | 141.4, C | — | 178.0, C | — | 131.9, C |
| 2 | — | 138.2, C | — | 143.4, C | — | 181.8, C | — | 123.3, C |
| 3 | — | 124.1, C | — | 124.4, C | — | 137.1a), C | — | 155.1, C |
| 4 | 7.13, s | 120.3, CH | 7.38, s | 126.3, CH | 7.24, s | 144.0, CH | 7.19, s | 110.5, CH |
| 4a | — | 130.1, C | — | 130.8, C | — | 137.3a), C | — | 137.0, C |
| 5 | 7.30, d (8.5) | 121.7, CH | 7.23, m | 119.0, CH | 6.88, d (7.5) | 122.8, CH | 7.28, d (8.0) | 119.3, CH |
| 6 | 7.14, dd (8.0, 8.5) | 123.4, CH | 7.23, m | 129.6, CH | 7.55, dd (7.5, 9.0) | 137.2, CH | 7.43, dd (8.0, 8.0) | 130.3, CH |
| 7 | 6.72, d (8.0) | 104.0, CH | 6.86, m | 110.6, CH | 7.02, d (9.0) | 114.6, CH | 6.80, d (8.0) | 109.5, CH |
| 8 | — | 154.9, C | — | 152.4, C | — | 163.0, C | — | 151.4, C |
| 8a | — | 114.5, C | — | 117.3, C | — | 118.7, C | — | 115.6, C |
| 9 | 3.86, s | 45.6, CH2 | 3.91, s | 46.3, CH2 | 2.45, dd (8.5, 14.5) | 39.4, CH2 | 3.54, d (16.5) | 40.0, CH2 |
| 2.67, dd (4.0, 14.5) | 3.58, d (16.5) | |||||||
| 10 | — | 206.8, C | — | 208.8, C | 4.05, m | 66.8, CH | — | 96.7, C |
| 11 | 2.22, s | 29.5, CH3 | 2.33, s | 30.3, CH3 | 1.26, d (6.0) | 23.7, CH3 | 1.83, s | 28.7, CH3 |
| 12 | — | — | — | — | — | — | — | 205.5, C |
| 13 | — | — | — | — | — | — | 2.73, s | 33.6, CH3 |
| 1-OMe | — | — | 4.06, s | 56.1, CH3 | — | — | — | — |
| 8-OMe | 4.06, s | 56.1, CH3 | — | — | 3.98, s | 56.4, CH3 | — | — |
| 1-OH | 9.12, s | — | — | — | — | — | — | — |
| 2-OH | 5.78, s | — | 6.76, s | — | — | — | — | — |
| 8-OH | — | — | 9.01, s | — | — | — | — | — |
a) Exchangeable.
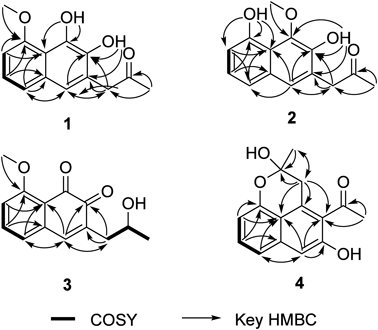
The molecular formula of 2 was established as C14H14O4, and its 1D NMR spectra were very similar to those of 1 (Table 1). In fact, the two-dimensional NMR analyses revealed that 2 comprises the identical tetrasubstituted naphthalene skeleton to that of 1 (Fig. 2). On the other hand, the HMBC correlations from two phenolic protons and a methoxy group and the downfield 13C resonance of the methoxy carbon (δC 62.5)10) strongly suggested that the methoxy group in 2 is attached to C1 (Fig. 2), and the chemical structure of 2 was completely determined (Fig. 1).
3 was isolated as yellow oil, and its molecular formula was identical to those of 1 and 2 (C14H14O4). The 13C-NMR spectrum of 3 also displayed 14 resonances, and the subsequent 1H-NMR and HMQC analyses revealed that it consisted of two carbonyl groups, four non-protonated aromatic carbons, four aromatic methines, one aliphatic oxymethine, two methyls, and one aliphatic methylene. Notably, the signal corresponding to a non-conjugated ketone disappeared, and a signal corresponding to an oxymethine (δC 66.8/δH 4.05) appeared instead. Therefore, the ketone group at the C10 position is likely to be reduced to an alcohol in 3, and this was confirmed by the COSY correlations among H9/H10/H11 (Fig. 2). It was also notable that only eight aromatic carbons were observed in the 13C-NMR spectra, along with two newly observed carbonyl signals (δC 178.0 and 181.8). Furthermore, no phenolic protons were observed in the 1H-NMR spectrum of 3, while the signals corresponding to four aromatic protons and methoxy group were observed. Therefore, 3 is likely to contain an ortho naphthoquinone chromophore derived from a 1,2-dihydroxy naphthalene unit. The COSY and HMBC correlations derived from H5, H6, and H7, and the methoxy protons indicated that 3 possessed the trisubstituted benzene ring A, identical to 1 (Fig. 2). In addition, the connection of C4/C4a was established by the HMBC correlations from H4 and H5, while the chemical bonds among C2, C3, C4, and C9 were constructed by the HMBC correlations from H4 and H9 (Fig. 2). To satisfy the molecular formula of 3, the remaining carbonyl carbon (C1, δC 178.0) should be present between C2 and C8a, and the chemical structure of 3 was fully established (Fig. 1). The optical rotation value of 3 was [α]25D −57.5° (c 0.033, MeOH), and the absolute configuration of C-10 was established as R with the modified Mosher’s method (Fig. S2).
The molecular formula of 4 was established as C15H14O4, indicating nine degrees of unsaturation. Consistently with the molecular formula, the 13C-NMR spectrum of 4 displayed 15 resonances, and the further edited-heteronuclear single quantum coherence (HSQC) analysis revealed that they consisted of one ketone, six non-protonated aromatic carbons, four aromatic methines, one hemiketal carbon, one aliphatic methylene, and two methyls. The presence of the trisubstituted benzene ring A was inferred from the COSY and HMBC correlations derived from H5, H6, and H7 (Fig. 2). In addition, the connections of C2/C12/C13 were inferred from the HMBC correlations from H13 (Fig. 2). The connectivities of C2/C3/C4/C4a were established by the HMBC correlations derived from the H4 signal, and the HMBC correlations from H9 to the aromatic carbons established the chemical bonds of C1/C2 and C1/C8a. Finally, the remaining C–C bonds (C1/C9/C10/C11) were constructed by the series of HMBC correlations derived from H9 and H11 (Fig. 2). Considering the chemical shifts in the 13C-NMR spectrum (Table 1), the unassigned three oxygen atoms must be adjacent to C3 (δC 155.1), C8 (δC 151.4), and C10 (δC 96.7). Furthermore, to satisfy the molecular formula of 3, it was most likely that C8 and C10 are connected through an ether bond to construct the six-membered hemiketal moiety (ring C in Fig. 1). Therefore, 4 was determined to be a naphthalene derivative (Fig. 1), which has not been reported as a natural product. Its optical rotation value was almost zero, [α]25D +3.6° (c 0.010, MeOH), indicating that 4 is likely to be racemic. This is reasonable, since the acetal moiety is formed nonenzymatically (Fig. S1).
1–3 were novel naphthalene or ortho naphthoquinone derivatives. Their chemical structures were closely related to juglomycin F11) and nocardiones12) (Fig. S3), but the C4 position of 1–3 were not oxidized unlike these metabolites. 4 is a tricyclic naphthalene derivative which has not been reported as natural product. Chemical structure of 4 is very similar to that of plant-derived metabolite xanthorrhoeol,13) while 4 contains the additional hydroxy group at C10 (Fig. S3).
Since phenolic (catecholic) compounds were reported to scavenge radical species and act as antioxidants,14) we evaluated the radical scavenging activities of the major components of 1–3 based on the 1,1-diphenyl-2-picrylhydrazyl (DPPH) assay.15) As expected from the presence of free phenolic OH, 1 and 2 displayed radical scavenging activity against DPPH, and the IC50 values of 1 and 2 were determined to be 15.6 and 12.8 µM, respectively. On the other hand, 3, lacking the free phenolic OH, did not show any radical scavenging activity against DPPH up to 100 µM. We also evaluated the antibacterial activities of 1–3, and only 3 displayed weak growth inhibitory activity against methicillin-sensitive Staphylococcus aureus (diameter of growth inhibitory zone: 8 mm, at a concentration of 10 µg per disk (6 mm)).
We searched the genome sequence of N. beijingensis for the biosynthetic genes for 1–4, and found one type II polyketide synthase (PKS) gene cluster encoding the enzymes possessing high identity with the erdacin biosynthetic enzymes16) (Table S1, Fig. S1). Due to the fact that there is only one type II PKS gene cluster in the genome and the analogy with the erdacin biosynthetic genes, it is likely responsible for the production of 1–4. Remarkably, the Blast Search with the IMG genome database (https://img.jgi.doe.gov) revealed that the focused putative biosynthetic gene clusters were widely distributed in 37 Nocardia and 2 Mycobacterium species (Fig. S4). Expectedly, N. anaemiae NBRC 100462, which also includes a putative beijinchrome-type gene cluster, produced juglomycin F (5).11) This data suggested that the beijinchrome-type gene cluster is also expressed in this strain.
In summary, we report the isolation of four novel antioxidant aromatic compounds, beijinchromes A–D (1–4) from N. beijingensis NBRC 16342. This is the first report on the secondary metabolites from N. beijingensis, and it opens a way toward understanding untapped pharmaceutical bioresources. The wide distribution of the putative biosynthetic gene clusters in Nocardia genomes implies their potential to produce the derivatives of 1–4 and explore the important biological functions of beijinchromes.
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (JSPS KAKENHI Grant Number JP16H06443, JP17H04763, and JP18K19139), JST/NSFC Strategic International Collaborative Research program Japan-China “Exploitation of the cryptic secondary metabolites from plant microbiome through biological interaction” (JPMJSC 1701), and JSPS Research Fellowships for Young Scientists (to S.H.). We also thank a Grant-in-Aid for the Cooperative Research Project from Institute of Natural Medicine, University of Toyama in 2017.
The authors declare no conflict of interest.
The online version of this article contains supplementary materials.