Experimental
General Procedure Pertaining to SynthesisAll reagents were commercially available with high purity grade. TLC was performed on precoated plates, TLC sheets silica 60 F254 (Merck, Darmstadt, Germany) or TLC sheets Chromatorex NH silica (Fuji Silysia Chemical, Kasugai, Japan). Chromatography was carried out on Silica Gel 60N (40–100 mesh) (Kanto Chemical, Tokyo, Japan) and NH silica gel Chromatorex (NH, 100–200 mesh) (Fuji Silysia Chemical). NMR spectra were recorded on a JEOL (Tokyo, Japan) JNM-AL300 (300 MHz) and Bruker (Billerica, MA, U.S.A.) Avance 600 (600 MHz). Chemical shifts were referenced to tetramethylsilane (TMS). Mass spectrum (FAB) and high-resolution mass spectra (HRMS) were recorded by a JEOL JMS-DX303. HRMS were recorded by using positive fast atom bombardment (FAB) with 3-nitrobenzyl alcohol (NBA) as the matrix. IR-red spectra were recorded on a JASCO (Tokyo, Japan) FT/IR-410. The samples were prepared as KBr discs or thin films between sodium chloride discs. Melting points were determined on a Yanaco (Kyoto, Japan) melting point apparatus and were uncorrected.
Synthesis of SN-4(Nps)22-({[2-(tert-Butylthio)ethyl]amino}methyl)-6-(diethoxymethyl)-N,N-dimethylpyridin-4-amine (2):Aldehyde 126) (1.6 g, 6.4 mmol) was dissolved in 45 mL of dist. MeOH, molecular sieves 4A and 2-(tert-butylthio)ethan-1-amine (2.7 g, 16 mmol) were added, and the mixture was stirred under an argon atmosphere for 25 h. Then, NaCNBH3 (1.2 g, 19 mmol) was added, and the mixture was stirred for 20 h. The mixture was filtered to remove molecular sieves, 10 mL of purified water was added, and the mixture was stirred for 30 min, and then the solvent was evaporated under reduced pressure. After adding 10 mL of purified water to the residue and extracting with CH2Cl2 (30 mL × 3), the organic layer was dried with MgSO4. The organic layer was concentrated under reduced pressure and purified by column chromatography (CH2Cl2 : MeOH = 20 : 1). 2 (2.0 g, 99%) was obtained as a brown oil. 1H-NMR (CDCl3) δ: 1.24 (6H, t, J = 7.03 Hz, CH3 ×2), 1.32 (9H, s, CH3 ×3), 2.73 (2H, t, J = 6.57, CH2), 2.84 (2H, t, J = 6.03 Hz, CH2), 3.01 (6H, s, CH3 ×2), 3.55–3.75 (4H, m, CH2 ×2), 3.87 (2H, s, CH2), 5.32 (1H, s, CH), 6.53 (1H, s, C5H2N (CH)), 6.71 (1H, s, C5H2N (CH)). 13C-NMR (CDCl3) δ: 15.1, 27.6, 30.9, 40.1, 42.6, 48.7, 63.1, 98.9, 103.7, 104.6, 151.4, 154.4, 157.4. FAB-MS (m/z): 370 (M + H)+.
4-(But-2-yn-1-yloxy)-N-[2-(tert-butylthio)ethyl]-N-{[6-(diethoxymethyl)-4-(dimethylamino)pyridin-2-yl]methyl}benzenesulfonamide (3):Amine 2 (2.0 g, 5.4 mmol) was dissolved in 18 mL of tetrahydrofuran (THF) and 0.98 mL of Et3N, 4-(but-2-yn-1-yloxy)benzenesulfonyl chloride27) (2.0 g, 8.1 mmol) dissolved in 7 mL of THF was added at 0 °C, and the mixture was stirred at room temperature for 11 h. Then, water was added, the mixture was stirred for 10 min. The solution was concentrated under reduced pressure and extracted with CH2Cl2. The extract was dried over MgSO4 and concentrated under reduced pressure. The residue was purified by color chromatography (hexane : AcOEt = 1 : 1) to afford 3 (2.0 g, 65%) as a yellow liquid. 1H-NMR (CDCl3) δ: 1.13 (15H, m, CH3 ×5), 1.81 (3H, s, CH3), 2.49 (2H, t, J = 7.9 Hz, CH2), 2.93 (6H, s, CH3 ×2), 3.20 (2H, t, J = 7.8 Hz, CH2), 3.54 (4H, q, J = 7.3 Hz, CH2 ×2), 4.30 (2H, s, CH2), 4.64 (2H, s, CH2), 5.17 (1H, s, CH), 6.56 (1H, s, C5H2N (CH)), 6.65 (1H, s, C5H2N (CH)), 6.98 (2H, d, J = 8.6 Hz, C6H4 (CH ×2)), 7.72 (2H, d, J = 8.6 Hz, C6H4 (CH ×2)). 13C-NMR (CDCl3) δ: 3.65, 15.2, 27.4, 31.0, 39.2, 42.5, 49.8, 54.8, 56.6, 62.3, 73.0, 84.7, 102.6, 103.2, 104.8, 115.1, 129.2, 131.5, 155.7, 156.0, 157.9, 161.0. IR (neat) 2973, 2314, 1604, 1500, 1442, 1338, 1230, 1153, 1110, 732, 1002, 917, 836 cm−1. High resolution (HR)MS (FAB) m/z Calcd for C29H44N3O5S2 (M + H)+ 578.2722. Found: 578.2725.
4-(But-2-yn-1-yloxy)-N-[2-(tert-butylthio)ethyl]-N-{[4-(dimethylamino)-6-formylpyridin-2-yl]methyl}benzenesulfonamide (4):Acetal 3 (0.30 g, 0.52 mmol) was dissolved in 3 mL of THF, 2M HCl was added at 0 °C, and the mixture was stirred at 50 °C for 3 h. The solution was neutralized with Et3N, concentrated under reduced pressure, extracted with CH2Cl2, dried with MgSO4 and concentrated under reduced pressure. The residue was purified by column chromatography (Hexane : AcOEt = 1 : 2) to afford 4 (0.18 g, 67%) as a yellow liquid. 1H-NMR (CDCl3) δ: 1.21 (9H, s, CH3 ×3), 1.88 (3H, s, CH3), 2.57 (2H, t, J = 8.1 Hz, CH2), 3.07 (6H, s, CH3 ×2), 3.33 (2H, t, J = 8.1 Hz, CH2), 4.45 (2H, s, CH2), 4.71 (2H, s, CH2), 6.89 (1H, s, C5H2N (CH)), 7.06 (2H, d, J = 8.8 Hz, C6H4 (CH ×2)), 7.08 (1H, s, C5H2N (CH)), 7.79 (2H, d, J = 8.8 Hz, C6H4 (CH ×2)), 9.85 (1H, s, CHO). 13C-NMR (CDCl3) δ: 3.70, 27.4, 30.98, 39.4, 42.6, 50.1, 54.5, 56.7, 84.8, 104.0, 108.5, 115.2, 129.2, 154.0, 155.5, 161.1, 194.2. IR (KBr) 2962, 2815, 2333, 1712, 1600, 1504, 1438, 1338, 1249, 1153, 998, 921, 840, 721 cm−1. HRMS (FAB) m/z Calcd for C25H33N3O4S2Na (M + Na)+ 526.1810. Found: 526.1830.
4-(But-2-yn-1-yloxy)-N-[2-(tert-butylthio)ethyl]-N-{[6-({[2-(tert-butylthio) ethyl]amino}methyl)-4-(dimethylamino)pyridin-2-yl]methyl}benzenesulfonamide (5):Aldehyde 4 (1.4 g, 2.8 mmol) was dissolved in 30 mL of dist. MeOH and added molecular sieves 3A, and the mixture was stirred for 20 min. Then 2-(tert-butylthio)ethan-1-amine was added and the mixture was stirred at 50 °C. for 24 h. NaCNBH3 (0.55 g, 8.8 mmol) was added, and the mixture was stirred at room temperature for 24 h. Molecular sieves 3A were removed by filtration, 10 mL of water was added, and the mixture was stirred at room temperature for 30 min. The solution was concentrated under reduced pressure and extracted with CH2Cl2. The extract was dried over MgSO4 and concentrated under reduced pressure. The residue was purified by column chromatography (CH2Cl2 : MeOH = 20 : 1) to afford 5 (1.1 g, 64%) as a brown liquid. 1H-NMR (CDCl3) δ: 1.21 (9H, s, CH3 ×3), 1.32 (9H, s, CH3 ×3), 1.88 (3H, s, CH3), 2.23 (1H, s, NH), 2.57 (2H, t, J = 8.1 Hz, CH2), 2.72 (2H, t, J = 6.6 Hz, CH2), 2.83 (2H, t, J = 6.6 Hz, CH2), 2.99 (6H, s, CH3 ×2), 3.28 (2H, t, J = 8.1 Hz, CH2), 3.74 (2H, s, CH2), 4.35 (2H, s, CH2), 4.71 (2H, s, CH2), 6.46 (1H, s, C5H2N (CH)), 6.54 (1H, s, C5H2N (CH)), 7.05 (2H, d, J = 8.8 Hz, C6H4 (CH ×2)), 7.79 (2H, d, J = 8.8 Hz, C6H4 (CH ×2)). 13C-NMR (CDCl3) δ: 3.72, 27.5, 28.7, 31.0, 39.3, 42.1, 42.5, 49.2, 49.8, 54.7, 54.9, 56.7, 73.4, 84.8, 103.7, 103.9, 115.1,129.2, 131.6, 155.7, 156.2, 161.0. IR (neat) 2958, 2348, 1600, 1500, 1450, 1338, 1226, 1153, 998, 917, 833, 732 cm−1. HRMS (FAB) m/z Calcd for C31H49N4O3S3 (M + H)+ 621.2967. Found: 621.2963.
4-(But-2-yn-1-yloxy)-N-({4-(dimethylamino)-6-[({2-[(2-nitrophenyl)dithio]ethyl}[(2-nitrophenyl)thio]amino)methyl]pyridin-2-yl}methyl)-N-{2-[(2-nitrophenyl) dithio]ethyl}benzenesulfonamide (6):Compound 5 (41 mg, 0.066 mmol) was dissolved in 0.7 mL of DMF and 1.5 mL of AcOH, 2-nitrobenzenesulfenyl chloride (Nps-Cl) (63 mg, 0.33 mmol) was added at 0 °C, and the mixture was stirred for 5 h. The resulting mixture was neutralized with sat. NaHCO3, and extracted with CH2Cl2. The extract was dried over MgSO4. and concentrated under reduced pressure. The residue was purified by column chromatography (Hexane : AcOEt = 1 : 1) to afforded 6 (45 mg, 70%) as a yellow solid. 1H-NMR (CDCl3) δ: 1.88 (3H, s, CH3), 2.81 (2H, t, J = 7.2 Hz, CH2), 2.92–2.97 (8H, m, CH3 ×2, CH2), 3.33–3.46 (4H, m, CH2 ×2), 4.19 (2H, s, CH2), 4.28 (2H, s, CH2), 4.71 (2H, s, CH2), 6.37 (1H, s, C5H2N (CH)), 6.46 (1H, s, C5H2N (CH)), 7.03 (2H, d, J = 8.9 Hz, C6H4 (CH ×2)), 7.27 (1H, m C6H4 (CH)), 7.30–7.33 (2H, m, C6H4 (CH ×2)), 7.61–7.64 (3H, m, C6H4 (CH)), 7.75 (2H, d, J = 8.9 Hz, C6H4 (CH)), 8.07 (1H, d, J = 12.0 Hz, C6H4 (CH)), 8.14 (1H, d, J = 8.2 Hz, C6H4 (CH)), 8.17 (1H, d, J = 8.2 Hz, C6H4 (CH)), 8.20–8.23 (2H, m, C6H4 (CH ×2)), 8.28 (1H, d, J = 8.2 Hz, C6H4 (CH)). 13C-NMR (CDCl3) δ: 3.7, 36.4, 36.8, 39.2, 47.9, 55.6, 56.8, 60.4, 64.4, 73.1, 76.8, 77.0,77.2, 104.6, 104.7, 114.9, 115.2, 124.9, 125.3, 125.9, 126.0, 126.1, 126.1, 126.2, 127.3, 129.4, 134.1, 137.4, 161.2. IR (KBr) 2341, 1600, 1511, 1446, 1338, 1153, 925, 848, 736 cm−1. HRMS(FAB) m/z Calcd for C41H42N7O9S6 (M + H)+ 968.1368. Found: 968.1352.
4-(But-2-yn-1-yloxy)-N-({4-(dimethylamino)-6-[({2-[(2-nitrophenyl)dithio]ethyl}amino)methyl]pyridin-2-yl}methyl)-N-{2-[(2-nitrophenyl)dithio]ethyl} benzenesulfonamide (SN-4(Nps)2):Compound 6 (40 mg, 0.041 mmol) was dissolve in a mixture of CH2Cl2 and MeOH (4.5 mL, 2 : 3), 3-methylindole (22 mg, 0.16 mmol) and 0.5 M HCl (0.2 mL) were added at 0 °C, and the mixture was stirred at room temperature for 6 h. The solution was concentrated under reduced pressure, neutralized with sat. NaHCO3, and extracted with CH2Cl2. The extract was concentrated under reduced pressure. The residue was purified by amino silica gel column chromatography (CH2Cl2 : AcOEt = 15 : 1) to afford a yellow solid SN-4(Nps)2 (18 mg, 54%). 1H-NMR (CDCl3) δ: 1.88 (3H, s, CH3), 2.80 (2H, t, CH2), 2.86–2.93 (10H, m, CH3 ×2, CH2 ×2), 3.40 (2H, t, J = 7.7 Hz, CH2), 3.64 (2H, s, CH2), 4.29 (2H, s, CH2), 4.71 (2H, s, CH2), 6.34 (1H, s, C5H2N (CH)), 6.47 (1H, s, C5H2N (CH)), 7.02 (2H, d, J = 8.9 Hz, C6H4 (CH ×2)), 7.33–7.35 (2H, m, C6H4 (CH)), 7.64–7.66 (2H, m, C6H4 (CH ×2)), 7.74 (2H, d, J = 8.9 Hz, C6H4 (CH ×2)), 8.14 (1H, d, J = 9.4 Hz, C6H4 (CH)), 8.25 (2H, d, J = 9.6 Hz, C6H4 (CH ×2)), 8.30 (1H, d, J = 9.4 Hz, C6H4 (CH)). 13C-NMR(CDCl3) δ: 3.72, 39.2, 47.7, 48.0, 54.7, 55.0, 56.8, 73.0, 84.9, 104.0, 115.2, 126.1, 126.2, 127.2, 127.4, 129.4, 130.8, 134.1, 137.3, 137.7, 145.5, 145.7, 155.6, 155.6, 161.2. IR (KBr) 3085, 2337, 1600, 1511, 1446, 1334, 1160, 917, 848, 744 cm−1. HRMS(FAB) m/z Calcd for C35H39N6O7S5 (M + H)+ 815.1484. Found: 815.1501. mp: 105 °C. Purity: 97.1% (See supplementary materials).
Cell Culture, Differentiation, and StimulationThe human monocytic cell line THP-1 was cultured in RPMI-1640 (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, MO, U.S.A.) and 55 µM 2-mercaptoethanol. The human astrocytoma cell line U251 MG was cultured in Dulbecco’s modified Eagle’s medium (DMEM) (FUJIFILM Wako Pure Chemical Corporation) supplemented with 10% FBS. All media were supplemented with 89 µg/mL streptomycin (Meiji Seika Pharma, Tokyo, Japan), and cells were incubated at 37 °C in a humidified atmosphere of 95% air and 5% CO2. The THP-1 cells were differentiated by incubation for 1 d with phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich) at 300 nM. The differentiated THP-1 cells were stimulated with lipopolysaccharide (LPS) (Sigma-Aldrich), and stimulation of the U251 MG cells was performed by ionomycin (LKT Laboratories, Saint Paul, MN, U.S.A.) or PMA (Sigma-Aldrich).
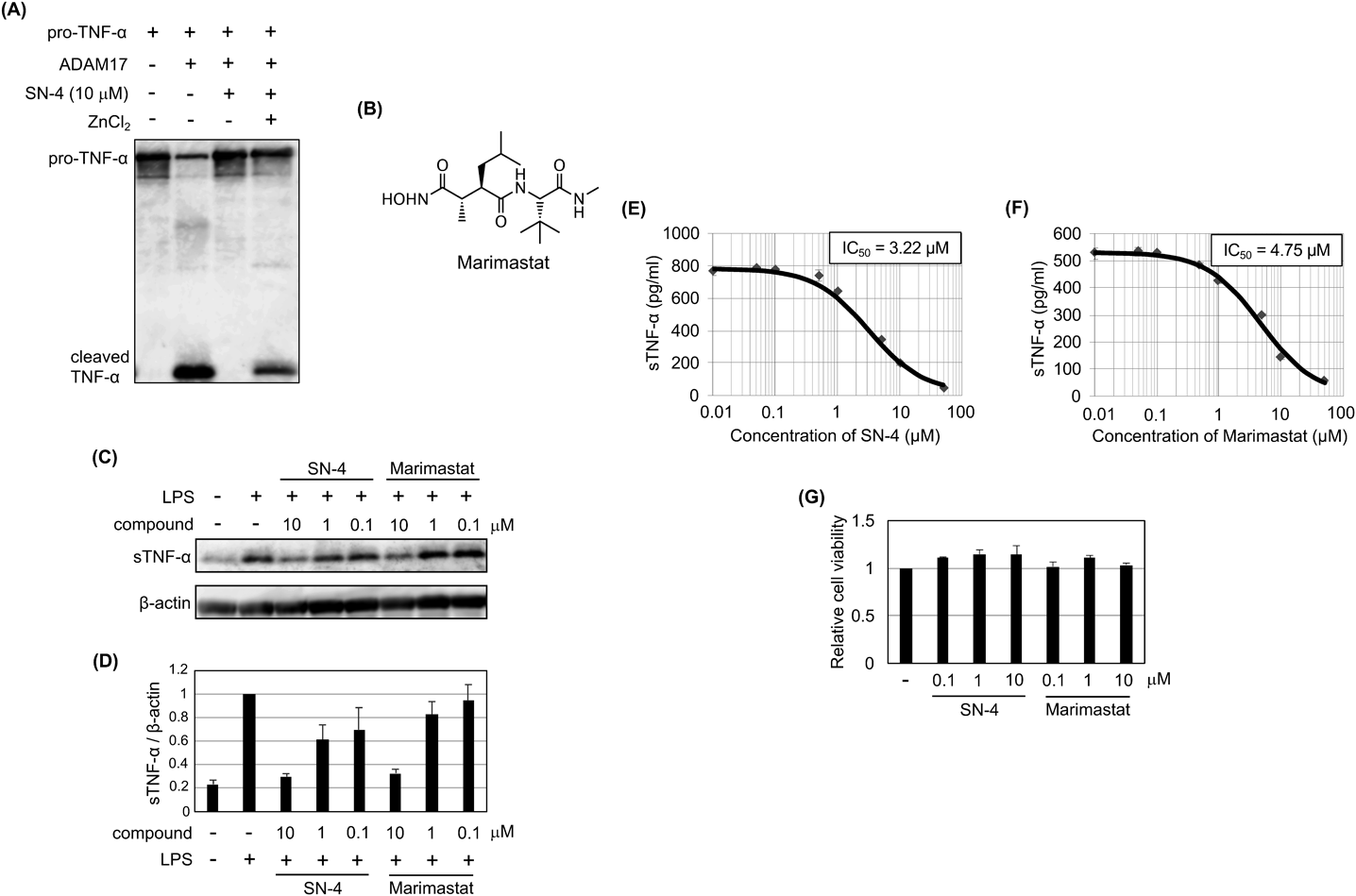
ChemicalsThe compound SN-4(Nps)2 was synthesized as described above. A control inhibitor marimastat was purchased from AnaSpec (Fremont, CA, U.S.A.). The SN-4(Nps)2 or marimastat was dissolved in dimethyl sulfoxide (DMSO) (FUJIFILM Wako Pure Chemical Corporation), and the solution was added to culture medium or in vitro reaction solution in a 1/100 volume.
MTT Assay and Analysis of Cell InvasionAmounts of viable cells were evaluated using MTT (Dojindo Molecular Technologies, Kumamoto, Japan) as previously described.37,38) Cell invasion was evaluated using Matrigel invasion chamber (BD Biosciences, Franklin Lakes, NJ, U.S.A.). U251 MG cells were seeded in upper transwell chamber (1 × 105 cells/well) and incubated for 1 d in presence of each compound. Noninvasive cells in upper surface of the membrane were removed by cotton tipped applicator, and invasive cells in lower surface of the membrane were stained with crystal violet (Nacalai Tesque, Kyoto, Japan) or their amounts were examined by MTT assay.
ImmunostainingU251 MG cells treated with compounds and PMA were fixed with 4% phosphate buffered saline (PBS) solution of paraformaldehyde (Tokyo Chemical, Tokyo, Japan) for 15 min at room temperature, and washed three times with PBS. The cells were incubated with PBS containing 5% normal goat serum (FUJIFILM Wako Pure Chemical Corporation) and 0.3% Triton X-100 (Nacalai Tesque) for 1 h. After removal of PBS, the cells were reacted with anti-CD44 monoclonal antibody BU52 (Ancell, Bayport, MN, U.S.A.) (1 : 400) in PBS containing 1% BSA (Nacalai Tesque) and 0.3% Triton X-100 overnight at 4 °C. The cells were then washed three times with PBS, reacted with goat anti-mouse immunoglobulin G (IgG) (H + L) Cross-Absorbed Secondary Antibody, Alexa Fluor 488 (Thermo Fisher Scientific, Waltham, MA, U.S.A.) at room temperature in the dark for 1 h. After washing three times with PBS, the cells were incubated with Hoechst 33342 (Dojindo Molecular Technologies) solution at RT for 15 min, followed by washing three times with PBS. Microscopic observation was performed using TCS SP5 confocal laser-scanning microscope (Leica, wetzlar, Germany).
Immunoblot Analysis and ELISAImmunoblot analysis was performed using cells lysed in PBS/Laemmli sample buffer (1 : 1) or cell culture supernatant as described previously.39) As an antibody, Human TNF-alpha Antibody (R&D Systems, Minneapolis, MN, U.S.A.) (1 : 500), anti-CD44 ICD (intracellular domain) Polyclonal antibody (TransGenic, Kobe, Japan) (1 : 500), or anti-β-actin clone AC-15 (Sigma-Aldrich) (1 : 1000) was used. Immunoreactivity was detected by using ImmunoStar LD (FUJIFILM Wako Pure Chemical Corporation). Intensity of bands was quantified by ImageJ (National Institute of health, Bethesda, MD, U.S.A.). ELISA was conducted by using Human TNF-α ELISA kit (Thermo Fisher Scientific).
In Vitro TNF-α Cleavage AssayMixture of Recombinant Human TACE/ADAM17 Protein (R&D Systems), SN-4(Nps)2 and 1,4-dithiothreitol (DTT) (FUJIFILM Wako Pure Chemical Corporation) with or without zinc chloride (FUJIFILM Wako Pure Chemical Corporation) in Tris–HCl buffer (50 mM, pH 8.0) was incubated at 37 °C for 30 min. Then Recombinant Pro-TNF-alpha Fusion Protein (R&D Systems) was added. The solution was incubated at 37 °C overnight followed by immunoblot analysis. The final concentrations are as follows. ADAM17, 100 µg/mL; SN-4(Nps)2, 10 µM; DTT, 30 µM; zinc chloride; 20 µM, Pro-TNF-α, 100 µg/mL.
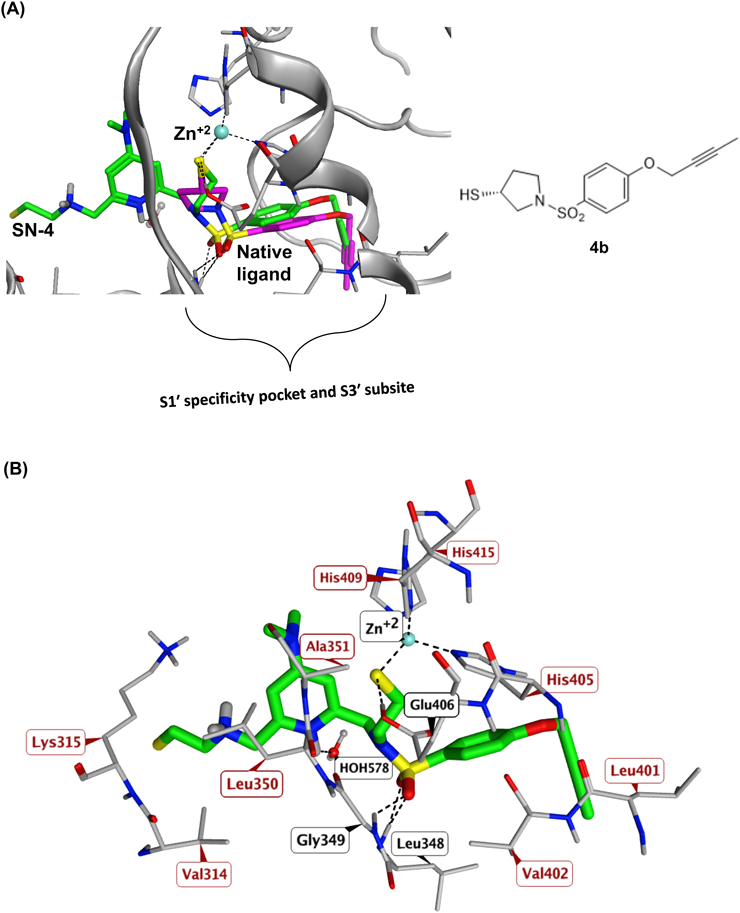
Molecular Docking AnalysisADAM-17 X-ray crystal structure co-crystalized with an Aryl-sulfonamide ligand (PDB code 2OI0)24) was retrieved from Protein Data Bank to be utilized as a model in the present study. The protein structure was prepared using QuickPrep module of MOE (Version 2019.0) (Chemical Computing Group, Montreal, Canada). Pocket water molecules and zinc ion were conserved. The docking study was conducted using the rigid-receptor method.36,40) The co-crystallized ligand was defined as the center of the binding site. All other options were left at their default values. One hundred docking positions were generated for each ligand. The generated docking positions were visualized using MOE.41)