2022 年 70 巻 3 号 p. 195-198
2022 年 70 巻 3 号 p. 195-198
We investigated similar compounds to ebselen and tideglusib, which exhibit strong activity against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), using Molecular ACCess System (MACCS) keys. Four candidate compounds were identified. One of them, phenyl-benzothiazol-3-one, showed coronavirus-specific 3C-like (3CL) protease inhibitory activity. The results indicated that a similarity score above 0.81 is a good indicator of activity for ebselen-and-tideglusib-like compounds. Subsequently, we simulated the ring-cleavage Michael reaction of ebselen at the Se center, which is responsible for its 3CL protease inhibitory activity, and determined the activation free energy of the reaction. The results showed that reaction simulation is a useful tool for estimating the activity of inhibitory compounds that undergo Michael addition reactions with the relevant cysteine S atom of 3CL proteases.
Since the end of 2019, coronavirus disease (COVID)-19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread worldwide. The spread of COVID-19 has caused numerous problems around the world, such as increases in the numbers of seriously ill patients and patient deaths, overwhelmed medical services, and postinfection sequelae. An mRNA vaccine for this infectious disease has been developed, and its administration is underway, but its effects cannot be fully confirmed yet. To date, remdesivir and dexamethasone have been approved as therapeutic agents for severely ill patients, and an antibody cocktail therapy (Ronapreve) has been approved and is being used as a therapeutic agent for mildly ill patients. However, to date, there is no treatment for asymptomatic individuals. In addition, mutations in the spike gene of the causative virus SARS-CoV-2 can occur, leading to ongoing concerns about the acquisition of resistance to current vaccines that target spikes. Accordingly, the development of effective preventive methods and therapeutic agents for COVID-19 is an urgent and ongoing challenge.
In the present study, we investigated research papers concerning COVID-19, which has been widespread since its outbreak in 2019; SARS, which was first identified in 2002; and 3C-like (3CL) proteases specific to coronavirus. This protease is the major viral proteinase that regulates the activity of the coronavirus replication complex; therefore, it is an attractive target for treatment.1) Information on nitrogen-containing organic compounds with inhibitory activity was extracted. There are numerous reports on the causal relationship between structural similarity and activity.2) Accordingly, we focused on compounds similar to ebselen and tideglusib based on the similar property principle that we have obtained by organic synthesis.3,4) Ebselen, a seleno organic compound, exhibits glutathione peroxidase-like activity. It is one of the promising synthetic antioxidants.5) It exhibits bioactivity by forming a seleno sulfide bond with the thiol group of the cysteine (Cys) residue of the protein. Studies on the conformation of the 3CL protease of SARS-CoV-2 and ebselen using molecular modeling have confirmed the fitting of ebselen to the active site of this enzyme.6) In addition, molecular dynamics simulations and LC/MS showed that ebselen is located in Cys145 of 3C-like protease in the catalytic cavity via a selenosulfide bond.7)
This report presents the 3CL protease inhibitory activities of these analogs and demonstrates potency prediction using the molecular modeling simulation method.
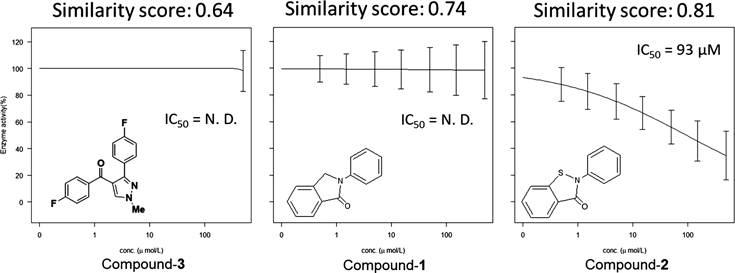
The published compounds considered in this study are ebselen, tideglusib, and PX-12, which are active against SARS-CoV-2 3CL protease2) and 45240, which is active against the 3CL protease of SARS-CoV,8) the causative SARS virus. The similarity between these chemical structures and those of the compounds we synthesized was evaluated using the Molecular ACCess System (MACCS) keys program. We focused on three compounds (compounds 1, 2,4) and 33)) which have a Tanimoto coefficient similarity score of 0.6 or higher (Fig. 1).

For the candidate compounds, activity tests were performed using 3CL protease assay kits (BPS Bioscience, CA, U.S.A.), and inhibition curves were plotted. Their IC50 values were calculated from the obtained inhibition curves. The 3CL protease inhibitory activities of compounds 1 and 3 are low (IC50 > 500 µM). However, compound 2, which has a similarity score of 0.81 (vs. ebselen) shows 3CL protease inhibitory activity (Fig. 2). This result suggests that a MACCS key similarity score of 0.81 or higher is an indicator of SARS-CoV-2 3CL protease inhibitory activity.
Therefore, based on candidate compounds 1 and 2, which have high similarity scores, the 3CL protease inhibitory activities of compounds in which the Se atom of ebselen is replaced with C or S and in which the phenyl group is replaced with hydrogen were measured. The results are shown in Fig. 3.
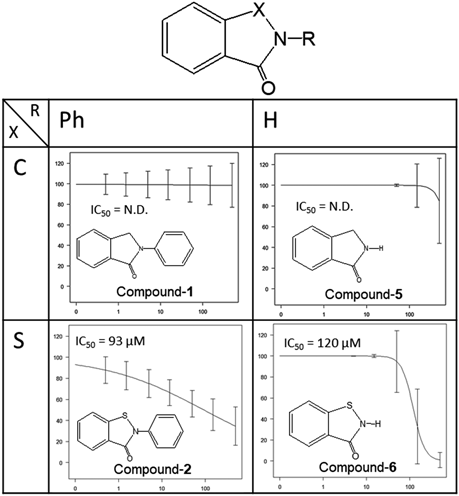
Thus, the compound in which X is a S atom and the R group is a phenyl ring has the highest 3CL protease inhibitory activity of the compounds considered.
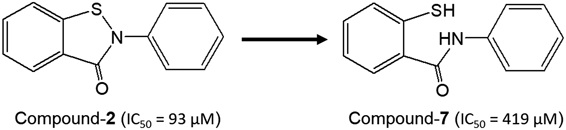
The activity of ebselen is based on a mechanism in which the Se atom undergoes an addition reaction with the S atom of a cysteine residue and the Se-containing five-membered ring opens.7) This mechanism has been demonstrated by X-ray crystal analysis of 3CL protease bound to ebselen as an inhibitor.2) The 3CL protease inhibitory activity of the ring-cleaved version of compound 2, which showed the strongest activity in the above activity test, was thus measured.
Interestingly, the ring-closed compound 2 (IC50 = 93 µM) shows higher inhibitory activity than the ring-opened compound 7 (IC50 = 419 µM; Fig. 4). Thus, it was concluded that the five-member-ring-closed compound is attacked by the S atom of a cysteine residue in the 3CL protease, and that a Michael reaction in which ring-opening and S addition happen simultaneously occurs.
Subsequently, we performed a molecular modeling simulation of this ring-opening reaction and calculated its predicted activation energy. We determined that the ease with which the ring-opening reaction proceeds is the source of the 3CL protease inhibitory activity.
First, a ring-cleavage reaction simulation was performed based on ebselen. As a result of modeling using density functional theory (DFT) at the ωB97XD/6-31+G(d) level and analyzing the data with the artificial force induced reaction (AFIR) method,9,10) we were able to propose the pathway of the reaction (Fig. 5).

Next, ring-cleavage simulations were also performed for Se(ebselen)-S and S(compound 2)-S, and the activation free energies were calculated. As a result, it was found that the reaction of ebselen has the lowest activation energy of 15.4 kcal/mol and that of candidate compound 2 has an activation energy of 24.3 kcal/mol. It was assumed that the candidate compound having a C atom instead of Se would not undergo this reaction (Table 1).
| Reactant | Type | Activate ΔG (kcal/mol) | 3CL Protease inhibition (IC50, µM) |
|---|---|---|---|
| Ebselen | Se-S | 15.4 | 0.67 |
| Compound 2 | S-S | 24.3 | 93.0 |
The 3CL protease inhibitory activity of compound 2 was reported to be 0.49 µM for IC50 in different evaluation systems.11) In our result compound 2 has a lower 3CL protease inhibitory activity than ebselen. It is suggested that the activation energy for the simulated five-membered-ring-cleavage reaction is an indicator of 3CL protease inhibitory activity for such compounds. This correlation may aid the design and preparation of derivatives of compound 2 as coronavirus-specific 3CL protease inhibitors.
The 3CL protease inhibitory activity of ebselen and its analogs is due to the Se atom of the five-membered ring. No activity is observed when the Se atom is replaced with a C or an O atom, but activity is observed when it is replaced with an S atom. We also established that the these activities are not due to reaction of the ring-opened state, but rather to the addition of S with the simultaneous opening of the five-membered ring. We also established that the activity of phenyl-benzothiazol-3-one (2) is lower than that of ebselen because the activation activity of the corresponding ring-opening reaction is higher for 2 than for ebselen. This demonstrates a new approach to activity prediction for such compounds.
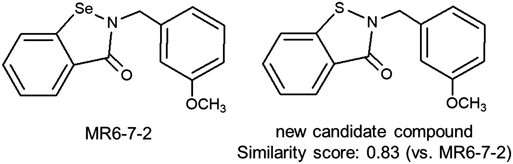
Accordingly, we have identified compound 2 as a lead compound. This compound has a simple chemical structure and it is not difficult to synthesize derivatives. Moreover, a recent study7) revealed that the ebselen derivative MR6-7-2 has potent 3CL protease inhibitory activity. Based on this structure, we propose a new candidate compound. The similarity score of this new candidate compound is calculated to 0.83 (Fig. 6). Thus, it may exhibit potent 3CL protease inhibitory activity.
The chemical structures of the test compounds were converted to simplified molecular input line entry system format. The Tanimoto coefficient was used to as a similarity score. The Rdkit program was used to calculate the similarity score by using MACCS keys.
Activation Free Energy Calculation for the Five-Membered-Ring-Cleavage Reaction Using the AFIR MethodA reaction paths for the five-membered ring cleavage were explored by the AFIR method,9,10) then these paths were improved using by the local updated planes (LUP) method. As a result of the LUP path, a candidate structures for the equilibrium and transition states were obtained respectively. Finally, all structures were fully optimized by the ωB97XD/6-31+G(d). The Gibbs free energies were evaluated at 298.15 K and 1 atm. All calculations were performed using the GRRM1412–16) combined with the Gaussian09.17)
SARS-CoV-2 3CL Protease InhibitionSARS-CoV-2-specific 3CL-protease-MBP-tagged assay kits were purchased from BPS Bioscience and used according to the manufacturer’s protocol. First, 4 ng/µL of 3CL protease-MBP-tagged enzyme in 30 µL of assay buffer was preincubated with 10 µL of the test compounds (final concentrations of 0, 0.5, 1.5, 5.0, 15, 150, and 500 µM in a final volume of 50 µL) for 30 min at 25 °C. The enzymatic reaction was initiated by adding 10 µL of substrate solution. The reaction mixture was incubated for 15 h at 25 °C. Wells with 1% dimethyl sulfoxide (DMSO), 4 ng/µL enzyme, and 50 mM substrate served as positive controls with no enzyme inhibition. Wells with GC376 (final concentrations of 2.5, 7.5, 25, 75, and 250 µM) served as the standard inhibitor and negative control. The fluorescence intensity of each reacted fraction was measured using an Infinite 200 Pro microplate-reading fluorimeter (TECAN, Switzerland). Based on the obtained fluorescence intensity, dose–response curves were created using the statistical software R to determine the IC50. Compounds 1, 2, 3, 4, and 7 were synthesized at Fukuyama University. Compounds 5 and 6 were obtained from Tokyo Chemical Industry (compound 5) and Kanto Chemical Co., Inc. (compound 6).
This research project was conducted as part of the 2020 Emergency Support: New Coronavirus Infection Control Subsidy program with support from the Nakatani Foundation. For more information on the Nakatani program, see https://www.nakatani-foundation.jp/achievements/covid-19_achievements_list/.
The authors declare no conflict of interest.