2019 Volume 66 Issue 5 Pages 431-441
2019 Volume 66 Issue 5 Pages 431-441
Forty-five pregnant women who underwent cesarean section, including 30 cases of gestational diabetes mellitus (GDM) and 15 normal pregnant women, were enrolled in this study to examine the differential expression of circular RNAs (circRNAs) in the placentas of women with GDM by RNA sequencing (RNA-seq) analysis. The differentially expressed circRNAs were analyzed bioinformatically using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment and circRNA-microRNA (miRNA) interaction prediction. Quantitative real-time polymerase chain reaction (qRT-PCR) was used to verify the results. A total of 8,321 circRNAs were identified in the human placenta, among which 46 were differentially expressed (fold change ≥2 and p < 0.05), including three that were upregulated and 43 that were downregulated. According to the GO and KEGG enrichment results, these circRNAs may be associated with vital biological processes, cellular components, molecular functions, and signaling pathways. In particular, KEGG analysis shown they may be involved in advanced glycation end products-receptor for advanced glycation end products (AGE-RAGE) signaling pathway in diabetic complications, indicating that these circRNAs might participate in the occurrence and pathogenesis of GDM. qRT-PCR verified that the expression of circ_5824, circ_3636, and circ_0395 was consistent with RNA-seq analysis; their expression levels were significantly lower in the GDM group than in the control group. The circRNA-miRNA interaction was analyzed according to the molecular sponge mechanism, and its potential function is discussed. These results shed light on future functional studies of circRNAs related to GDM.
THE PRESENT STUDY suggests that circRNAs are associated with the occurrence and development of GDM. Our results may open a new chapter in the study of GDM.
Gestational diabetes mellitus (GDM), also known as glucose intolerance with onset or first recognition during pregnancy [1], can cause severe maternal and neonatal complications, such as increased risk of preeclampsia, macrosomia, depression, and stillbirth. It is particularly noteworthy that uncontrolled GDM has long-term adverse effects on mothers and children, such as susceptibility to obesity and metabolic syndrome [2]. Because of its high incidence (affecting 3–9% of pregnancies [3]), GDM has attracted the attention of prenatal medical experts.
GDM is a heterogeneous disorder in which pregnancy can make recessive diabetes dominant (make pregnant women with no previous diabetes have GDM) or aggravate the condition of a woman with preexisting diabetes. The hallmark of GDM is increased insulin resistance [4]. Pancreatic beta cells are no longer able to compensate for the increased insulin resistance during pregnancy. However, the precise mechanisms underlying GDM remain unknown. On the other hand, Ilekis et al. [5] suggested that the occurrence of adverse pregnancy outcomes associated with GDM could be related to placental issues. As the main channel of energy transfer between mother and fetus, it should play an important role in the abnormal metabolism of GDM. Placental lactogen, prolactin, and estradiol also seem to contribute to the development of insulin resistance during pregnancy, among which cortisol and progesterone are the main culprits. Other studies have focused on the placental microenvironment, including inflammatory factors, genes, and proteins [6-8]. The recent discovery of small molecules with potential regulatory effects, such as microRNAs (miRNAs) [9, 10] and long non-coding RNAs (lncRNAs) [11, 12], may reveal the essence of GDM more accurately.
Circular RNAs (circRNAs) are a special type of non-coding RNA with characteristics of evolutionary conservation, structural stability, and tissue specificity [13, 14]. Because of these biological features, circRNAs have attracted wide attention in recent years. They play important roles in tumor development and nervous system diseases [15-17]. circRNAs act as miRNA sponges and affect the expression of downstream genes [18, 19]. Some reports have shown that circRNAs are also present in the human placenta and may be related to the occurrence of pregnancy complications [20]. circRNAs may also play a role in the pathogenesis of diabetes and thus serve as novel molecular targets for clinical therapy [21, 22]. For example, Zhao et al. reported that hsa_circ_0054633 presented a certain diagnostic capability for pre-diabetes and type 2 diabetes mellitus, and high glucose exposure profoundly altered circRNA expression in endothelial cells [23]. However, studies on the relationship between circRNAs and GDM are lacking.
In the present study, we examined the differential expression of circRNAs in placentas from women with GDM by RNA sequencing (RNA-seq) and preliminarily investigated their biological functions via bioinformatics analysis. We hope to clarify the relationship between circRNAs and the occurrence and pathogenesis of GDM.
A total of 45 pregnant women who underwent cesarean section from August 2016 to June 2017, including 30 cases of GDM and 15 normal pregnant women, were enrolled in this study. Their baseline characteristics are shown in Table 1. The diagnosis of GDM was made by an oral glucose tolerance test (75 g) during the second trimester (24–28 weeks of gestation). All pregnant women with multiple gestations, infection, other pregnancy complications, congenital or chromosomal abnormalities of the fetus, or a family history of diabetes were excluded. The study was approved and reviewed by the ethics committee of Changzhou Women and Children Health Hospital (Changzhou, China, Approval No: CZFY20160103). Informed consent was obtained prior to cesarean section.
| Characteristics | GDM (N = 30) | Control (N = 15) | p value |
|---|---|---|---|
| Age (year) | 32.57 | 31.47 | 0.39 |
| Fasting glucose (mmol/L) | 5.09 | 4.35 | 0.03 |
| 2 h postgrandial glucose (mmol/L) | 6.86 | 5.28 | 0.001 |
| OGTT 1 h (mmol/L) | 10.27 | 7.32 | <0.01 |
| OGTT 2 h (mmol/L) | 8.98 | 6.06 | <0.01 |
| HbA1C (%) | 5.18 | 4.85 | 0.13 |
| BMI at delivery (kg/m2) | 28.97 | 27.27 | 0.21 |
| Pre-pregnancy BMI (kg/m2) | 22.12 | 21.34 | 0.22 |
| Birth weight (g) | 3,445.67 | 3,362.85 | 0.61 |
| Neonatal sex (male/female) | 0.87 | 0.88 | 0.55 |
Placentas were obtained within 15 min after cesarean section. Placental fragments were collected in the middle of the initial placental depth. The decidual layer, chorionic surface, and membranes were removed. All placental samples were washed with saline and stored at −80°C following the addition of 1 mL Trizol (Invitrogen, Carlsbad, CA, USA). Three cases and three paired controls were chosen for RNA-seq analysis.
RNA extraction and sequencingTotal RNA was extracted from the placenta tissues using the mirVana miRNA Isolation Kit (Ambion, Inc., Foster City, CA, USA) following the manufacturer’s protocol. RNA integrity was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Samples with an RNA integrity number ≥7 were subjected to subsequent analyses. Libraries were constructed using TruSeq Stranded Total RNA with Ribo-Zero Gold according to the manufacturer’s instructions. Libraries were then sequenced on the Illumina sequencing platform (HiSeq 2500) and 150 bp/125 bp paired-end reads were generated.
Identification and quantification of human circRNAscircRNAs were predicted by CIRCexplore2 [24] and compared with those in circBase (http://www.circbase.org/). DESeq [25] software was used to standardize the number of junction reads of each sample. A fold change (FC) >2 and p < 0.05 were considered to indicate significant differences.
Functional enrichment analysis and circRNA-miRNA associationsThe differentially expressed circRNA genes were subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. GO enrichment analysis was based on three aspects: biological process (BP), cellular component (CC), and molecular function (MF). circRNA-targeted miRNAs were identified and predicted by miRanda [26, 27]. The circRNA-miRNA interaction network was constructed based on the functional annotation of the miRNA target genes.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysisWe randomly selected 10 circRNAs from the RNA-seq results to verify by qRT-PCR. RNase R-treated RNAs were diluted with water and used as a PCR template. cDNAs were obtained using a Reverse Transcription Kit (M-MLV; Promega, Madison, WI, USA). SYBR Master Mix (TaKaRa Bio Inc., Kusatsu, Japan) was used to examine the expression of circRNAs according to the manual. All primers were designed and synthesized by Ribo Bio (Guangzhou, China). Amplification was performed on an ABI 7500 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) under the following conditions: denaturation at 95°C for 10 min, followed by 38 cycles of amplification at 95°C for 10 s and 60°C for 1 min. The relative expression levels of the circRNA genes were calculated using the 2−ΔΔCt method. GAPDH was employed as the internal reference to normalize the expression levels of the target genes.
Statistical analysisStatistical analyses were performed using SPSS 19 software. The t-test was used to analyze data between two groups. P < 0.05 was considered statistically significant.
A total of 8,321 circRNAs were indentified in human placentas from GDM and normal control pregnant women. A total of 7,804 circRNAs had already been reported in circBase, whereas 517 were newly discovered, among which 46 were differentially expressed in the placenta tissues of GDM women (FC > 2 and p < 0.05), three that were upregulated and 43 that were downregulated. Their related information is shown in Table 2. A heatmap (Fig. 1A) and volcano plot (Fig. 1B) reveal the differential expression profiles of circRNAs between GDM women and the control group.
| ID | CircBase name | region | gene symbol | chr | log2FC | p | regulation |
|---|---|---|---|---|---|---|---|
| circ_0395 | NA | exonic | PAPPA2 | 1 | –4.69 | 0.02 | down |
| circ_1698 | hsa_circ_0058092 | exonic | FN1 | 2 | –6.46 | 0.05 | down |
| circ_1840 | hsa_circ_0081006 | exonic | KRIT1 | 7 | –6.68 | 0.04 | down |
| circ_1926 | hsa_circ_0000155 | exonic | DCAF6 | 1 | –6.74 | 0.04 | down |
| circ_2308 | hsa_circ_0120939 | exonic | EXOC6B | 2 | –6.61 | 0.05 | down |
| circ_2372 | hsa_circ_0006260 | exonic | SLC41A2 | 12 | –7.90 | 0.01 | down |
| circ_2415 | hsa_circ_0007430 | exonic | NRDC | 1 | 7.17 | 0.03 | up |
| circ_2523 | hsa_circ_0000857 | exonic | ZNF236 | 18 | –7.19 | 0.03 | down |
| circ_3003 | hsa_circ_0005362 | exonic | PHC3 | 3 | –6.65 | 0.04 | down |
| circ_3223 | hsa_circ_0002466 | exonic | TTBK2 | 15 | –7.69 | 0.02 | down |
| circ_3636 | NA | exonic | ADAM12 | 10 | –5.90 | 0.03 | down |
| circ_3798 | hsa_circ_0005243 | exonic | TMEM184B | 22 | –8.17 | 0.04 | down |
| circ_3869 | hsa_circ_0001578 | exonic | RANBP9 | 6 | –7.85 | 0.01 | down |
| circ_3993 | hsa_circ_0008192 | exonic | PTBP3 | 9 | –6.44 | 0.05 | down |
| circ_4046 | hsa_circ_0088249 | exonic | PAPPA | 9 | –6.55 | 0.05 | down |
| circ_4390 | hsa_circ_0006670 | exonic | SIPA1L3 | 19 | –6.87 | 0.03 | down |
| circ_4524 | hsa_circ_0006380 | exonic | TCF12 | 15 | –6.46 | 0.05 | down |
| circ_4718 | hsa_circ_0005029 | exonic | EPT1 | 2 | –6.65 | 0.04 | down |
| circ_4792 | hsa_circ_0000417 | exonic | CPSF6 | 12 | –6.87 | 0.03 | down |
| circ_4802 | hsa_circ_0002702 | exonic | RUSC2 | 9 | –7.40 | 0.04 | down |
| circ_5036 | hsa_circ_0009049 | exonic | PLPP3 | 1 | 6.90 | 0.03 | up |
| circ_5124 | hsa_circ_0028319 | exonic | TMEM116 | 12 | –6.49 | 0.05 | down |
| circ_520 | hsa_circ_0004919 | exonic | CARF | 2 | –6.69 | 0.04 | down |
| circ_5754 | NA | exonic | LOC100507487 | 4 | –6.45 | 0.05 | down |
| circ_5824 | hsa_circ_0005243 | exonic | TMEM184B | 22 | –2.37 | 0.04 | down |
| circ_6525 | hsa_circ_0134318 | exonic | GLI3 | 7 | –6.63 | 0.04 | down |
| circ_6998 | hsa_circ_0013218 | exonic | DNTTIP2 | 1 | –7.01 | 0.03 | down |
| circ_7167 | hsa_circ_0002226 | exonic | ETFA | 15 | –6.77 | 0.04 | down |
| circ_7224 | hsa_circ_0002795 | exonic | SPAST | 2 | –6.99 | 0.03 | down |
| circ_730 | hsa_circ_0042170 | exonic | NCOR1 | 17 | –6.52 | 0.04 | down |
| circ_7360 | hsa_circ_0002634 | exonic | ATXN7 | 3 | –6.44 | 0.05 | down |
| circ_7367 | hsa_circ_0003218 | exonic | BMPR2 | 2 | –7.88 | 0.01 | down |
| circ_7402 | hsa_circ_0126389 | exonic | SLC30A9 | 4 | –6.69 | 0.04 | down |
| circ_7466 | hsa_circ_0125310 | exonic | LARP1B | 4 | 6.76 | 0.04 | up |
| circ_7540 | hsa_circ_0091581 | exonic | GPC3 | X | –6.37 | 0.05 | down |
| circ_7687 | hsa_circ_0008667 | exonic | ADAMTS6 | 5 | –7.19 | 0.03 | down |
| circ_780 | hsa_circ_0002814 | exonic | HERC2 | 15 | –6.54 | 0.04 | down |
| circ_7965 | hsa_circ_0025641 | exonic | RASSF8 | 12 | –6.95 | 0.03 | down |
| circ_8068 | hsa_circ_0017310 | exonic | CNST | 1 | –6.73 | 0.04 | down |
| circ_8086 | hsa_circ_0008234 | exonic | FOXP1 | 3 | –7.11 | 0.03 | down |
| circ_8122 | NA | splicing | LIMS2 | 2 | –6.64 | 0.04 | down |
| circ_8133 | hsa_circ_0000139 | exonic | GON4L | 1 | –8.05 | 0.01 | down |
| circ_8210 | hsa_circ_0002968 | exonic | MAPK8 | 10 | –7.34 | 0.02 | down |
| circ_8271 | hsa_circ_0091206 | exonic | PCDH11X | X | –7.10 | 0.03 | down |
| circ_967 | hsa_circ_0035472 | exonic | RNF111 | 15 | –7.25 | 0.05 | down |
| circ_986 | NA | exonic | PAPPA2 | 1 | –6.70 | 0.04 | down |
Note: NA, not available; chr, chromosome.
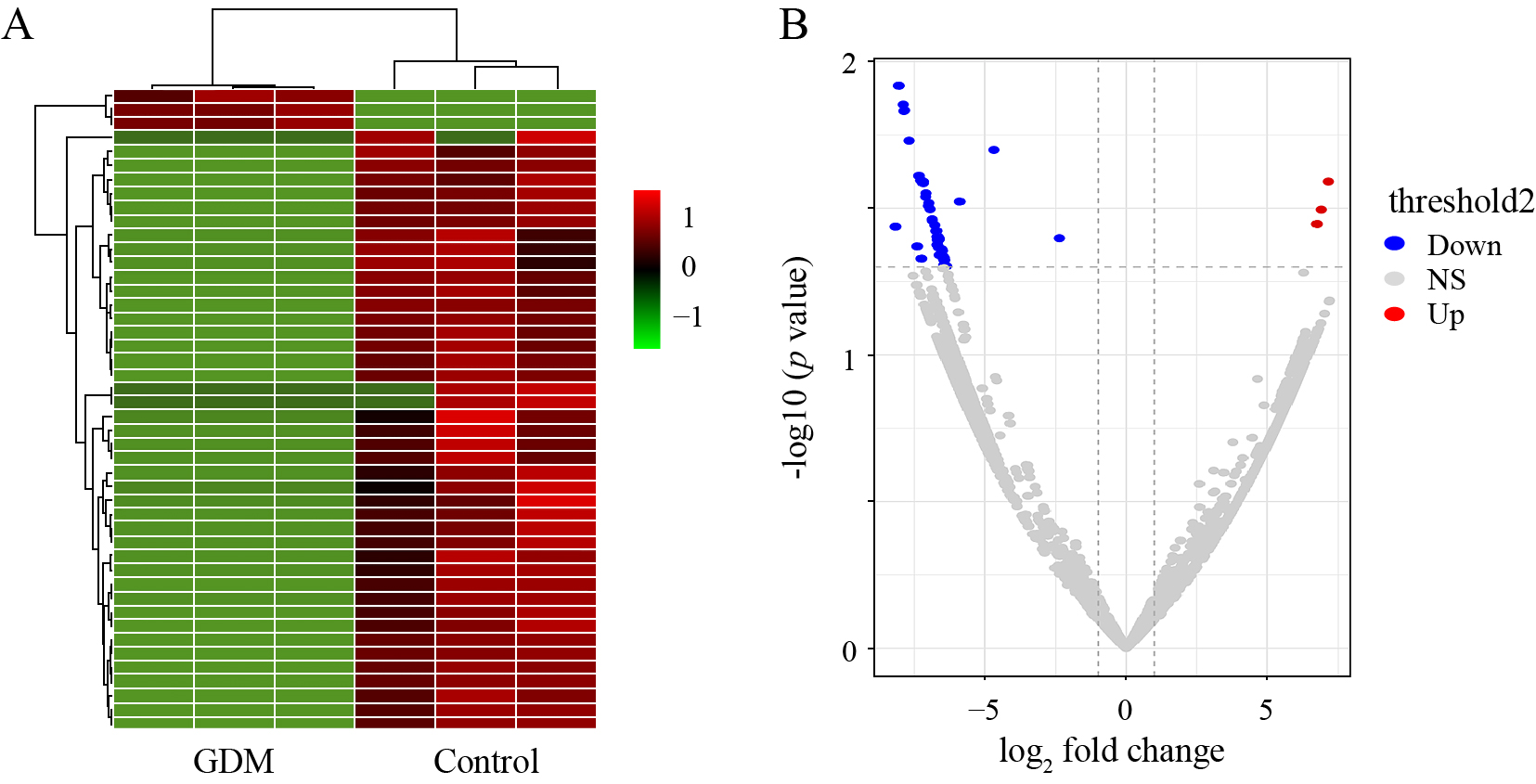
Prediction and identification of circRNAs expressed in the placentas of women with GDM
A. Expression profiles of the circRNAs are displayed in a heatmap. Each column represents a sample and each row represents a circRNA. High expression is indicated in red and low expression is indicated in green. B. Differentially expressed circRNAs are displayed in volcano plots. Gray dots indicate circRNAs with no significant difference. Red dots indicate significantly upregulated circRNAs, whereas blue dots indicate significantly downregulated circRNAs. NS, not significant.
The differentially expressed circRNA genes were analyzed by GO (Fig. 2A, B, C) and KEGG (Fig. 2D) enrichment. Based on the results, these differentially expressed circRNAs may be associated with GO functional annotation of biological processes (e.g., smoothened signaling pathway), cellular components (e.g., nuclear speck, transcriptional repressor complex), and molecular function (e.g., small ubiquitin-like modifier [SUMO] binding, ubiquitin−like protein binding). According to KEGG analysis, the host genes of these differentially expressed circRNAs are associated with focal adhesion, shigellosis, bacterial invasion of epithelial cells, endocrine resistance, and the advanced glycation end products-receptor for advanced glycation end products (AGE−RAGE) signaling pathway in diabetic complications. In particular, the AGE/RAGE pathway plays an important role in a variety of diabetic complications [28] and is associated with adverse outcomes in GDM [29].
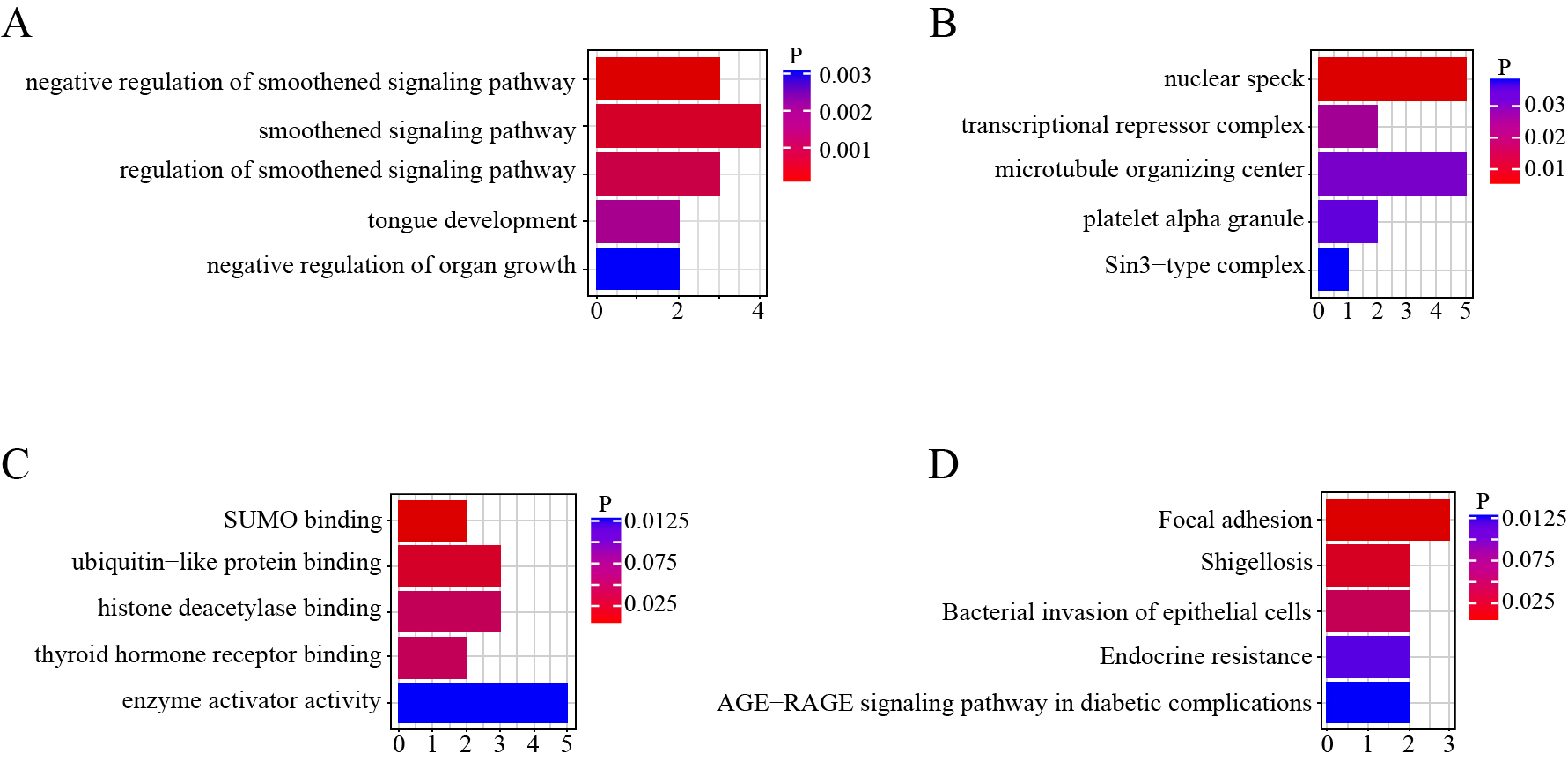
GO and KEGG enrichment terms of differentially expressed circRNA transcript genes
A. Top five classes of biological process (BP) enrichment terms. B. Top five classes of molecular function (MF) enrichment terms. C. Top five classes of cellular component (CC) enrichment terms. D. Top five classes of KEGG pathway terms.
To validate the RNA-seq results, 10 circRNAs were randomly subjected to qRT-PCR analysis, among which the results of three circRNAs (circ_5824, circ_3636, and circ_0395) were consistent with RNA-seq data. Compared with the control group, their expression in the GDM group was significantly reduced (Fig. 3A). Their primer sequences are shown in Table 3. Images of the PCR products of these three circRNAs on 1.5% agarose gels did not show obvious primer dimers or non-specific PCR products (Fig. 3B). All bands of the GDM group were less bright than those in the control group, indicating their differential expression.
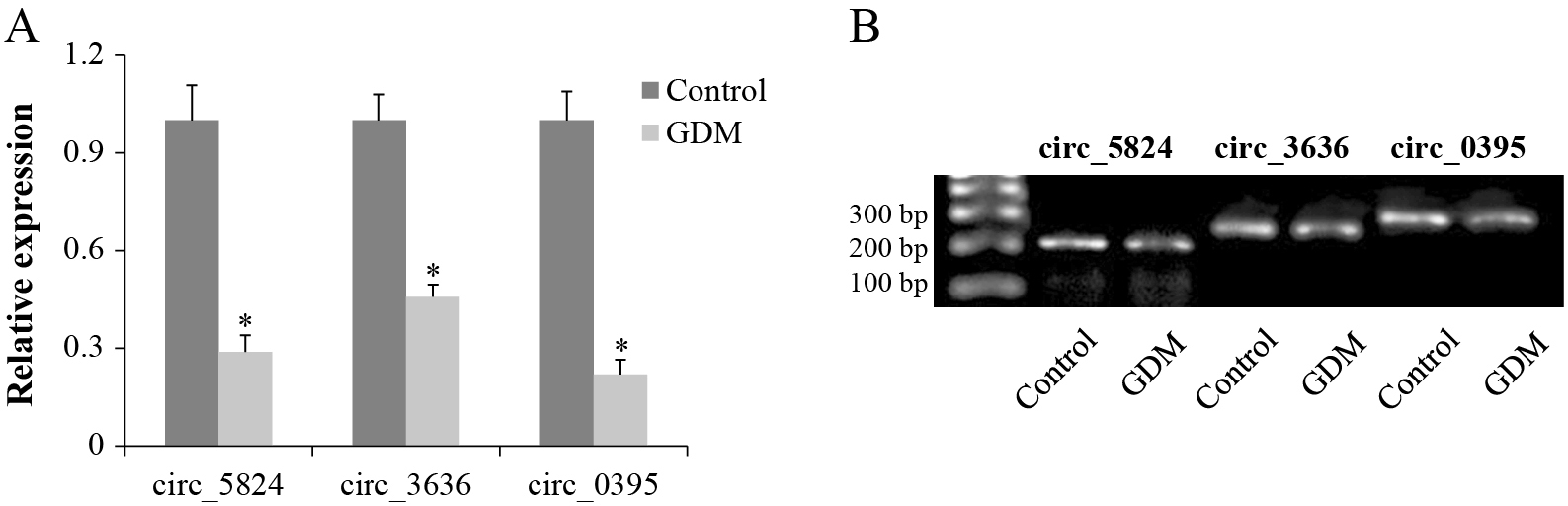
qRT-PCR verification of differentially expressed circRNAs
A. The expression of circ_5824, circ_3636, and circ_0395 was confirmed by qRT-PCR, which was consistent with the sequencing results. * p < 0.05. B. Images of PCR products of the three circRNAs on a 1.5% agarose gel.
| Gene | 5'-3' | 3'-5' |
|---|---|---|
| circ_5824 | CACCGGACAGGCATCTAGTGA | CAGTGTTGCAGGCTCTTTGA |
| circ_0395 | AGACAGGAATTTGGGTACATC | GAGTGCCATCCACATACAGG |
| circ_3636 | GTGCTATGGTGCTCTGTCTA | TGAGTGAGCCGAGTTGTTCT |
| GAPDH | TGACTTCAACAGCGACACCCA | CACCCTGTTGCTGTAGCCAAA |
As target molecules of miRNAs, the interaction analysis of circRNA-miRNA can help explore the function and mechanism of circRNAs. Three circRNA (circ_5824, circ_3636, and circ_0395)-targeted miRNAs were identified and their potential functions were elucidated according to the miRNA target genes. The miRNAs that interact with these three circRNAs, as predicted using miRanda software, are shown in Table 4. circRNA_0395 was targeted by 88 miRNAs. The top three total scores of these miRNAs were hsa-miR-8485, hsa-miR-3135b and hsa-miR-1273g-3p, whereas circRNA_3636 and circRNA_5824 had three and eight miRNA binding sites, respectively. Furthermore, a network of circRNA-miRNA-mRNA interactions was established based on these three circRNAs and their target miRNAs (Fig. 4).
| Transcript | miRNA | Total score | Total energy | miRNA length | Position |
|---|---|---|---|---|---|
| circRNA_0395 | hsa-miR-197-3p | 163 | –30.67 | 22 | 16149 |
| hsa-miR-143-5p | 171 | –30.17 | 22 | 6098 | |
| hsa-miR-363-5p | 173 | –32.57 | 22 | 21029 | |
| hsa-miR-328-5p | 156 | –30.21 | 23 | 29421 | |
| hsa-miR-328-3p | 171 | –33.9 | 22 | 15185 | |
| hsa-miR-504-3p | 490 | –91.63 | 21 | 17820 6102 37955 | |
| hsa-miR-619-5p | 192 | –42.14 | 22 | 5885 | |
| hsa-miR-33b-3p | 163 | –31.13 | 22 | 723 | |
| hsa-miR-762 | 152 | –31.86 | 22 | 4010 | |
| hsa-miR-877-3p | 173 | –33.32 | 21 | 37405 | |
| hsa-miR-937-5p | 164 | –32.56 | 20 | 1045 | |
| hsa-miR-939-3p | 161 | –32.92 | 21 | 32279 | |
| hsa-miR-1226-3p | 173 | –39.96 | 22 | 2418 | |
| hsa-miR-1207-5p | 325 | –61.96 | 21 | 45023 52033 | |
| hsa-miR-1285-3p | 183 | –33.2 | 22 | 45215 | |
| hsa-miR-1303 | 184 | –34.28 | 22 | 31482 | |
| hsa-miR-1304-3p | 167 | –30.42 | 22 | 17797 | |
| hsa-miR-1254 | 167 | –30.39 | 24 | 29504 | |
| hsa-miR-1273a | 195 | –40.51 | 25 | 20806 | |
| hsa-miR-1911-5p | 174 | –31.86 | 23 | 27861 | |
| hsa-miR-1912 | 167 | –30.6 | 22 | 3402 | |
| hsa-miR-1913 | 154 | –32.71 | 22 | 57642 | |
| hsa-miR-1972 | 176 | –34.62 | 22 | 21085 | |
| hsa-miR-1976 | 162 | –30.12 | 20 | 30368 | |
| hsa-miR-2276-5p | 164 | –34.09 | 22 | 44308 | |
| hsa-miR-3127-3p | 175 | –34.76 | 22 | 3440 | |
| hsa-miR-3137 | 154 | –30.13 | 24 | 29438 | |
| hsa-miR-3151-5p | 154 | –30.84 | 21 | 4010 | |
| hsa-miR-3184-3p | 175 | –31.45 | 23 | 35510 | |
| hsa-miR-3192-5p | 168 | –31.82 | 23 | 12012 | |
| hsa-miR-3200-5p | 180 | –31.15 | 22 | 58686 | |
| hsa-miR-4254 | 172 | –33.9 | 23 | 6254 | |
| hsa-miR-4269 | 169 | –30.86 | 21 | 22629 | |
| hsa-miR-3619-5p | 161 | –30.17 | 22 | 15775 | |
| hsa-miR-3135b | 633 | –134.88 | 22 | 6104 35487 37957 31448 | |
| hsa-miR-4518 | 159 | –30.96 | 26 | 41381 | |
| hsa-miR-4640-3p | 161 | –30.85 | 22 | 22760 | |
| hsa-miR-4644 | 183 | –31.99 | 23 | 29307 | |
| hsa-miR-4651 | 157 | –30.53 | 20 | 45812 | |
| hsa-miR-4656 | 164 | –30.76 | 23 | 29419 | |
| hsa-miR-4685-3p | 174 | –36.05 | 22 | 18357 | |
| hsa-miR-4687-5p | 165 | –33.59 | 22 | 44117 | |
| hsa-miR-4722-5p | 163 | –30.19 | 23 | 58192 | |
| hsa-miR-4728-5p | 166 | –30.49 | 23 | 45800 | |
| hsa-miR-4741 | 167 | –32.11 | 23 | 254 | |
| hsa-miR-4758-5p | 155 | –34.04 | 23 | 870 | |
| hsa-miR-4763-5p | 153 | –30.28 | 21 | 22772 | |
| hsa-miR-4763-3p | 174 | –36.51 | 24 | 52031 | |
| hsa-miR-4436b-5p | 162 | –32.29 | 22 | 8141 | |
| hsa-miR-5090 | 162 | –30.65 | 23 | 9284 | |
| hsa-miR-5095 | 187 | –41.97 | 21 | 5879 | |
| hsa-miR-1273g-3p | 548 | –112.25 | 21 | 29358 20828 8006 | |
| hsa-miR-5096 | 179 | –32.37 | 21 | 5957 | |
| hsa-miR-5187-5p | 176 | –30.51 | 22 | 31084 | |
| hsa-miR-5189-5p | 168 | –31.45 | 24 | 27993 | |
| hsa-miR-5196-3p | 157 | –36.54 | 21 | 302 | |
| hsa-miR-6089 | 340 | –84.82 | 24 | 32436 21040 | |
| hsa-miR-6727-5p | 157 | –30.15 | 23 | 9285 | |
| hsa-miR-6734-5p | 167 | –32.06 | 23 | 15550 | |
| hsa-miR-6734-3p | 173 | –30.38 | 23 | 38422 | |
| hsa-miR-6751-5p | 177 | –32.76 | 23 | 34482 | |
| hsa-miR-6756-5p | 533 | –118.76 | 23 | 4014 45811 56027 | |
| hsa-miR-6764-5p | 160 | –31.16 | 22 | 32 | |
| hsa-miR-6771-5p | 320 | –65.79 | 22 | 12050 20896 | |
| hsa-miR-6776-3p | 168 | –31.44 | 23 | 22795 | |
| hsa-miR-6777-3p | 166 | –32.42 | 20 | 30373 | |
| hsa-miR-6782-5p | 161 | –30.39 | 25 | 4011 | |
| hsa-miR-6787-3p | 166 | –31.64 | 22 | 17799 | |
| hsa-miR-6793-5p | 175 | –34.09 | 22 | 41435 | |
| hsa-miR-6797-5p | 165 | –30.74 | 25 | 29034 | |
| hsa-miR-6799-3p | 159 | –31.14 | 23 | 905 | |
| hsa-miR-6803-5p | 160 | –33.98 | 22 | 6836 | |
| hsa-miR-6810-5p | 165 | –30.76 | 23 | 9210 | |
| hsa-miR-6810-3p | 171 | –39.89 | 23 | 57643 | |
| hsa-miR-6812-5p | 328 | –66.88 | 25 | 45808 56018 | |
| hsa-miR-6780b-5p | 172 | –31.94 | 23 | 3663 | |
| hsa-miR-6846-5p | 162 | –32.59 | 22 | 549 | |
| hsa-miR-6849-5p | 171 | –30.56 | 23 | 23141 | |
| hsa-miR-6856-5p | 173 | –31.35 | 24 | 31975 | |
| hsa-miR-6884-3p | 179 | –38.62 | 23 | 37374 | |
| hsa-miR-7108-3p | 164 | –31.3 | 20 | 8900 | |
| hsa-miR-7111-3p | 347 | –66.25 | 22 | 38374 38427 | |
| hsa-miR-7160-3p | 157 | –33.98 | 21 | 44310 | |
| hsa-miR-1273h-5p | 334 | –65.67 | 21 | 12016 29392 | |
| hsa-miR-7851-3p | 176 | –32 | 22 | 12018 | |
| hsa-miR-8085 | 171 | –30.52 | 21 | 12693 | |
| hsa-miR-8089 | 160 | –32.08 | 24 | 45819 | |
| hsa-miR-8485 | 928 | –184.2 | 21 | 37704 44413 37720 44429 44383 | |
| circRNA_3636 | hsa-miR-6734-5p | 158 | –32.46 | 23 | 111 |
| hsa-miR-6852-5p | 162 | –31.45 | 21 | 73 | |
| hsa-miR-7155-5p | 167 | –30.87 | 19 | 78 | |
| circRNA_5824 | hsa-miR-331-3p | 154 | –33.14 | 21 | 287 |
| hsa-miR-486-3p | 162 | –30.4 | 21 | 340 | |
| hsa-miR-4486 | 170 | –34.58 | 17 | 111 | |
| hsa-miR-4649-5p | 153 | –30.89 | 24 | 100 | |
| hsa-miR-4786-3p | 168 | –31.26 | 22 | 194 | |
| hsa-miR-6165 | 156 | –34.53 | 19 | 26 | |
| hsa-miR-6729-5p | 160 | –32.59 | 22 | 105 | |
| hsa-miR-8073 | 176 | –33.56 | 22 | 34 |
A total of 99 miRNAs could be combined with these 3 circRNAs. Total Score: the cumulative prediction score. The higher the value is, the more accurate. Total Energy: the accumulative complementary pair matches the free energy. The smaller the energy is, the more reliable.

circRNA-miRNA-mRNA interaction network
The circRNA-miRNA-mRNA network consists of three circRNAs (blue), 21 miRNAs (red) and 120 disease-related genes (green).
We successfully discovered 46 differentially expressed circRNAs in the placentas of women with GDM via RNA-seq analysis and confirmed the results using qRT-PCR. Their biological functions were predicted by bioinformatics analysis. These results suggest that the differentially expressed circRNAs are associated with the occurrence and development of GDM.
The roles of several small molecules in the occurrence of GDM have recently attracted our attention, namely, miRNAs and lncRNAs. Thus far, more than 600 miRNAs expressed in the human placenta have been reported [30]. Human placenta tissue exhibits specific miRNA expression in a time-dependent manner during pregnancy and is reflected in the maternal plasma. Some placental miRNAs are dysregulated in plasma and are involved in GDM [9]. As a newly discovered non-coding RNA, circRNAs have gained increasing attention from researchers. Several functional circRNAs that act as competitive endogenous RNAs by effectively adsorbing miRNAs and regulating their target genes were recently identified [31, 32]. To date, there have been few reports on the relationship between circRNAs and pregnancy. Maass et al. detected 63 circRNAs in the human placenta. By functional prediction, they reported that some circRNAs may be related to pregnancy complications, such as early onset preeclampsia, fetal growth restriction, and infection during pregnancy [20]. In this study, we identified 46 differentially expressed circRNAs in the placentas of women with GDM and preliminarily discussed the possible mechanisms of their participation in GDM. Yan et al. [33] recently reported differentially expressed circRNAs in placenta tissues from patients with GDM. Their circRNA expression profile differed from that described herein, which could be due to the different prediction tools used (different algorithms have different sensitivities and accuracy rates; dramatic differences between the algorithms were observed specifically regarding the highly expressed circRNAs and the circRNAs derived from proximal splice sites) [34]. Different regions and populations, as well as database sequencing systems, will also cause differences.
Among the differentially expressed circRNAs, the qRT-PCR results of three circRNAs (circ_5824, circ_3636 and circ_0395) were consistent with RNA-seq analysis. circRNA_0395 attracted our attention; it was significantly decreased in the placentas of women with GDM and overlaps with the PAPPA2 (pregnancy-associated plasma protein A 2) gene, which encodes a pregnancy-related protein. PAPPA2 can be used to predict macrosomia at birth in GDM pregnancies [35] and is related to metabolic diseases beyond total adiposity [36]. Thus, it was important to examine the interactions between circRNAs and miRNAs that could play a key role in the occurrence and development of GDM. Accumulating evidence has indicated that circRNAs have a series of important biological functions, acting as miRNA sponges [16, 37]. Bioinformatics analysis revealed that circRNA_0395 is targeted by 88 types of miRNAs. We then performed a systematic review of the literature to examine these miRNAs more closely. miRNA-1273g-3p piqued our interest. miR-1273g-3p, a member of the miR-1273 family, was first identified as an miRNA in 2011 and can bind to 1,074 genes [30]. Guo et al. [38] reported that miR-1273g-3p participates in acute glucose fluctuation and is an important factor that leads to endothelial dysfunction and autophagy. It is also involved in the progression of several complications caused by diabetes by modulating the autophagy-lysosome pathway and could serve as a new target for disease therapy [39]. However, there has been no report on the role of miRNA-1273g-3p in pregnancy. Thus, without direct evidence that circRNA_0395 is targeted by miRNA-1273g-3p, further research was needed to confirm their relationship.
In conclusion, our study provides a preliminary landscape of the differential expression of circRNAs that may be involved in the occurrence and pathogenesis of GDM. The current study also provides new insight into the molecular mechanism of GDM.
Huiyan Wang, Guangtong She, Wenbai Zhou and Bin Yu carried out the assays and participated in designing the study. Huiyan Wang, Guangtong She, Kezhuo Liu, Jun Miao carried out clinical consultation. Guangtong She, Wenbai Zhou and Bin Yu carried out sample collection, laboratory tests and performed the statistical analysis. Guangtong She and Bin Yu conceived the study, participated in its design and coordination and draft the manuscript.
We thank all of the project participants for their contributions.
The authors declare that they have no competing interests.
The study design and protocol were reviewed and approved by the ethics committee of Changzhou Maternity and Child Health Care Hospital affiliated to Nanjing Medical University (Approval No: CZFY20160103).
This work was supported by grants from Jiangsu province health and family planning commission (F201439, QNRC305), Changzhou health and family planning commission (ZD201412), Changzhou municipal bureau of science and technology (CJ20159055).