2024 Volume 71 Issue 7 Pages 675-686
2024 Volume 71 Issue 7 Pages 675-686
Endothelial-to-mesenchymal transition (EndMT) is a pivotal event in diabetic retinopathy (DR). This study explored the role of circRNA zinc finger protein 532 (circZNF532) in regulating EndMT in DR progression. Human retinal microvascular endothelial cells (HRMECs) were exposed to high glucose (HG) to induce the DR cell model. Actinomycin D-treated HRMECs were used to confirm the mRNA stability of phosphoinositide-3 kinase catalytic subunit δ (PIK3CD). The interaction between TATA-box-binding protein-associated factor 15 (TAF15) and circZNF532/PIK3CD was subsequently analyzed using RNA immunoprecipitation (RIP), RNA pull-down. It was found that HG treatment accelerated EndMT process, facilitated cell migration and angiogenesis, and enhanced PIK3CD and p-AKT levels in HRMECs, whereas si-circZNF532 transfection neutralized these effects. Further data showed that circZNF532 recruited TAF15 to stabilize PIK3CD, thus elevating PIK3CD expression. Following rescue experiments suggested that PIK3CD overexpression partially negated the inhibitory effect of circZNF532 silencing on EndMT, migration, and angiogenesis of HG-treated HRMECs. In conclusion, our results suggest that circZNF532 recruits TAF15 to stabilize PIK3CD, thereby facilitating EndMT in DR.
DIABETIC RETINOPATHY (DR) is a microvascular complication of diabetes that is caused by retinal endothelial dysfunction and may lead to vision loss [1, 2]. It is estimated that the DR population worldwide will increase to 578 million in 2030 and approximately 700 million by 2045 [3]. The presence and progression of DR is associated with a significant increase in medical costs [4]. Unfortunately, current treatments against DR target late-stage of the disease may not effective in dealing with early changes. Endothelial-to-mesenchymal transition (EndMT) refers to the process whereby endothelial cells lose their typical characteristics and transform into mesenchymal cells in various pathological conditions [5]. During the pathogenesis of DR, hyperglycemia induces the retinal endothelial cells (RECs) to undergo EndMT [6]. RECs exposed to high glucose (HG) show upregulation of mesenchymal markers [7]. EndMT is involved in the formation of fibrous epiretinal membranes in DR, which is an important event in DR [8]. However, the underlying mechanisms regulating EndMT in DR have not been fully elucidated. Despite remarkable advances in treatment, DR is still a major cause of vision loss [9]. Hence, elucidating the pathological mechanism of DR related to EndMT is imperative to develop novel therapeutic strategies for DR.
Circular RNAs (circRNAs) are a new class of endogenous noncoding RNAs with regulatory functions and single-chain covalent closed structures [10, 11]. They are widely involved in the regulation of gene expression and can be used as potential therapeutic targets and biomarkers for a variety of diseases[12, 13]. Several studies have demonstrated the involvement of circRNAs in the regulation of various biological activities, including cell invasion, migration, and vascularization [14, 15]. Some studies have suggested the involvement of circRNAs in the regulation of EndMT in several human diseases [16-19]. Moreover, circRNAs have been shown to be crucial regulators of DR progression. For instance, circRNA_0084043 was found to aggravate HG-treated ARPE-19 cell injury by facilitating oxidative stress and inflammation [20]. circCOL1A2 was shown to enhance VEGF release and contribute to angiogenesis in DR [21]. CircRNA zinc finger protein 532 (circZNF532) consists of many isomers, in which hsa_circ_0047818 is located at chr18: 56601664-56606853. A previous study demonstrated the upregulation of circZNF532 in HG-induced human retinal microvascular endothelial cells (HRMECs), and the downregulation of circZNF532 restrained cell proliferation, migration, and angiogenesis [22]. Nonetheless, the potential impact of circZNF532 on EndMT in DR and its mechanism have not been clarified, underlining the need for further investigation.
Phosphoinositide-3 kinase catalytic subunit δ (PIK3CD or PI3Kδ) is a key mediator of cell growth, and its expression is markedly enhanced in HG-treated HRMECs [23]. Studies have revealed that PIK3CD facilitates EndMT and HRMEC viability by activating the AKT pathway [24, 25]. As previously described, some circRNAs can bind to RNA-binding protein to form complexes, which in turn regulate downstream gene transcription, mRNA stability, and translation [26]. TATA box binding protein (TBP)-associated factor 15 (TAF15 or TAFII68) is an RNA-binding protein that affects gene expression by directly binding to mRNAs and stabilizing them. For example, a previous study suggested that circDNAJC11 binds to TAF15 to accelerate breast cancer development by stabilizing MAPK6 mRNA and activating the MAPK signaling pathway [27]. According to StarBase prediction, circZNF532 and PIK3CD have potential binding sites for TAF15. However, the relationship between these three genes and the mechanism by which they regulate EndMT in DR remains unclear.
We speculated that circZNF532 recruits TAF15 to increase PIK3CD mRNA stability and promote EndMT during DR. In this study, we tested this hypothesis in an in vitro model of HG-triggered HRMECs. Our findings may provide new insights for developing a novel treatment method for DR.
HRMECs were obtained from Cell Systems Corporation (UK) and grown in a basic endothelial cell culture medium (5 mM glucose) containing 10% FBS (Gibco, Grand Island, NY, USA) and 1% penicillin/streptomycin (Gibco). 293T cells were acquired from Procell (Wuhan, China) and maintained in DMEM (Gibco) comprising 10% FBS (Gibco) and 1% penicillin/streptomycin (Gibco). HRMECs and 293T cells were grown in an incubator at 37°C in 5% CO2.
Establishment of a HG-induced cell modelHRMECs (5 × 105 cells/mL) were plated in 12-well plates. After fasting for 24 h, HRMECs were stimulated with the indicated doses of glucose, including HG (25 mmol/L D-glucose), normal glucose (control, 5 mmol/L D-glucose), and osmotic glucose (25 mmol/L L-glucose). The cells were grown in these conditioned media for 48 h [28].
Cell transfection and treatmentThe si-circZNF532 (target sequence: GATAGGAGAACCAAAAAGACT), si-TAF15 (target sequence: GAATTATCAAGACAAATAAGA), and negative control si-NC (target sequence: GTTCTCCGAACGTGTCACGT) were designed by RiboBio Co. Ltd. (Guangzhou, China). HRMECs (5 × 105 cells/mL) were seeded into 12-well plates. The next day, when approximately 80% confluence was reached, cells were transfected with si-circZNF532, si-TAF15, or si-NC using Lipofectamine2000 (Invitrogen, Carlsbad, CA, USA). Following transfection for 48 h, cells were starved for 24 h and then stimulated with different concentrations of glucose as mentioned above for 48 h. For actinomycin D treatment, after glucose stimulation, 5 μg/mL actinomycin D was added to cells and stimulated for 0 h, 2 h, and 4 h, respectively.
Lentivirus infectionThe lentivirus containing the circZNF532 overexpressing vector (lenti-circZNF532), TAF15 overexpressing vector (lenti-TAF15), PIK3CD overexpressing vector (lenti-PIK3CD), and control vector (lenti-vector) were obtained from Sangon Biotech Co. Ltd. (Shanghai, China). HRMECs (5 × 105 cells/mL) were seeded into 12-well plates for 12 h. Subsequently, 20 μL lentivirus at a viral titer of 1 × 108 TU/mL diluted in 1 mL DMEM was used to infect HRMECs.
Quantitative real-time PCR (qRT-PCR)HRMECs were harvested for total RNA isolation using TRIzol (Invitrogen) according to standard guidelines. The cDNA was reverse transcribed, followed by qRT-PCR assay using a 7,500 real-time PCR System (Applied Biosystems, Carlsbad, CA, USA) with SYBR Green master mix (Thermo Fisher, Waltham, MA, USA). circZNF532 expression and PIK3CD mRNA level were normalized to β-actin and analyzed using the 2–ΔΔCt method. The primer sequences are as follows: circZNF532 (F): TTTAAGTCTGCCCCAAGCAC, circZNF532 (R): AGGGCAGGTGTAGGGAGATT; PIK3CD (F): AAGGAGGAGAATCAGAGCGTT, PIK3CD (R): GAAGAGCGGCTCATACTGGG; GAPDH (F): CCAGGTGGTCTCCTCTGA, GAPDH (R): GCTGTAGCCAAATCGTTGT.
Western blot analysisProtein extraction from HRMECs was performed, and the protein concentration was examined using a BCA protein kit (Beyotime, Shanghai, China). A total of 30 μg protein was used for electrophoresis via 10% SDS-PAGE, followed by transfer to a PVDF membrane (Millipore, Billerica, MA, USA). The membranes were blocked in 5% BSA for 2 h and then incubated overnight at 4°C with primary antibodies, including anti-VE-Cadherin (1:500, V1514, Sigma-Aldrich, USA), anti-CD31(1:10,000, ab76533, Abcam, UK), anti-vimentin (1:500, SAB4503081, Sigma-Aldrich), anti-α-SMA (1:500, abs130621, Absin, China), anti-PIK3CD (1:2000, ab109006), anti-AKT (1:10,000, ab179463), anti-p-AKT (1:800, ab38449), anti-TAF15 (1:2000, ab133760) and anti-β-actin (1:5,000, ab8227). The membranes were then washed with TBST and incubated with HRP-labeled secondary antibodies (1:3,000, ab205718; 1:3,000, ab205719) for 1.5 h. After a final wash with TBST, the bands were visualized using enhanced chemiluminescence (ECL) reagents (Beyotime). The gray value was analyzed to assess relative protein levels using Image J software (NIH, Bethesda, MD, USA).
Immunofluorescent stainingHRMECs were fixed with 4% paraformaldehyde, permeabilized with 0.3% Triton X-100, and blocked with 5% normal goat serum. Next, HRMECs were incubated overnight at 4°C with primary antibodies anti-VE-Cadherin (1 μg/mL, ab33168) and anti-α-SMA (1:300, ab124964). After washing with cold PBS, HRMECs were incubated with secondary antibody goat anti-rabbit IgG (1:500, ab150077) for 2 h. Nuclei were stained with DAPI solution (Beyotime) for 5 min in the dark. Cells were visualized and photographed using fluorescence microscopy (Olympus, Tokyo, Japan).
Dual-luciferase reporter assayA wild-type or mutated fragment of PIK3CD was inserted into the psi-Check2 plasmid (PIK3CD-wt and PIK3CD-mut). These plasmids were cotransfected into 293T cells with the circZNF532-overexpressing vector (oe-circZNF532) or control vector (oe-NC) using Lipofectamine2000 (Invitrogen). After 48 h, the relative luciferase activity was detected using dual-luciferase reporter assays (E1910, Promega, WI, USA).
RNA-binding protein immunoprecipitation (RIP)This assay was conducted using a Magna RIPTM Kit (Millipore) to determine the binding between TAF15 and circZNF532 or PIK3CD. In brief, the cell lysates were immunoprecipitated with protein A/G agarose beads coupled to anti-TAF15 or anti-IgG antibodies at 4°C overnight. The complexes were washed and digested, and the abundance of circZNF532 and PIK3CD in the immunoprecipitants was assessed by qRT-PCR.
RNA pull-down assayAn RNA-protein Pull-Down Kit (Thermo Fisher) was used for the assay. In brief, the HRMECs were harvested and lysed, and the cell lysates were treated with biotinylated circZNF532 or PIK3CD containing streptavidin magnetic beads, which could capture the proteins interacting with circZNF532 or PIK3CD. After washing, the protein–RNA complex bound to the beads was eluted and the TAF15 protein level was analyzed by Western blot.
Transwell assayAfter corresponding transfection and treatment, a suspension of HRMECs at a density of 1 × 106 cells/mL was prepared. Then, 200 μL cell suspension was placed in the upper chamber of a 24-well Transwell plate (8 μm pore size, Corning, USA). For the lower chamber, a volume of 500 μL medium consisting of 10% FBS was added. The plate was then placed at 37°C with 5% CO2 for 24 h. Next, the medium and cells in the inside upper chamber were removed, and cells in the outside bottom of the upper chamber were fixed with 4% paraformaldehyde for 10 min, followed by staining with 0.5% crystal violet for 15 min. The migrated cells were observed and counted under an inverted light microscope (Olympus Corporation).
Matrigel tube formation assayFollowing the indicated transfection and stimulation, HRMECs were resuspended in serum-free DMEM, and 1.5 × 103 cells/100 μL were inoculated in 96-well plates precoated with solidified Matrigel (Millipore). After culture at 37°C for 10 h, tube formation was visualized using an inverted microscope (Olympus Corporation).
Statistical analysisData were presented as mean ± standard deviation, and between-group differences were assessed using Student’s t-test or one-way analysis of variance followed by Tukey’s post-hoc test. Data analyses were performed using SPSS 19.0 software (IBM Corp. USA); p < 0.05 was considered indicative of statistical significance.
First, circZNF532 was silenced in HRMECs by transfection of si-circZNF532, followed by stimulation with HG to induce a DR model in vitro. As shown in Fig. 1A, circZNF532 expression was increased after HG exposure and then decreased after transfection of si-circZNF532, suggesting the effective HG exposure and siRNA transfection. EndMT is characterized by reduced expression of endothelial markers (VE-Cadherin and CD31) and increased expression of mesenchymal markers (Vimentin and α-SMA) [29]. Based on the Western blot results, HG treatment induced downregulation of VE-Cadherin and CD31, and upregulated Vimentin and α-SMA levels in HRMECs, whereas si-circZNF532 transfection neutralized the effect induced by HG (Fig. 1B). Immunofluorescence staining data showed that VE-Cadherin protein was decreased in HG conditions and α-SMA protein was increased in the HG group. However, transfection with si-circZNF532 counteracted the decline of VE-Cadherin and elevation of α-SMA induced by HG (Fig. 1C). The Transwell assay results validated the increased number of migrated cells upon HG stimulation compared with the control or osmotic group, whereas si-circZNF532 transfection abrogated this promoting effect (Fig. 1D). In addition, HG stimulation facilitated HRVEC angiogenesis, and circZNF532 silencing reversed the angiogenic effect of HG exposure (Fig. 1E). These data suggest that circZNF532 promoted EndMT, migration, and angiogenesis of HRMECs upon HG stimulation.

Effect of circZNF532 silencing on EndMT, migration, and angiogenesis of HG-stimulated HRMECs. HRMECs were transfected with si-circZNF532 or si-NC followed by HG (25 mmol/L D-glucose) treatment for 48 h. Cells exposed to 5 mmol/L D-glucose or 25 mmol/L L-glucose served as the control or osmotic (osm) group, respectively. (A) Detection of si-circZNF532 transfection efficiency; (B) Western blot analysis of VE-Cadherin, CD31, Vimentin, and α-SMA; (C) Immunofluorescent staining for localization and expressions of VE-Cadherin and α-SMA; (D) Representative images of HRVEC migration obtained by Transwell assay. Quantitative data of the migrated cell number; (E) The Matrigel tube formation assay was used to assess the angiogenic ability of HRMECs. Statistical data of the total tube length. Each experiment was repeated at least three times. * p < 0.05, ** p < 0.01, *** p < 0.001.
To identify the downstream target of circZNF532, we searched previous reports and identified 12 genes that play important regulatory roles in EndMT of DR, including METTL3, SPRED2, Nrf2, AGK, miR-205-5p, miR-34a-5p, PKC, PIK3CD, Sirt1, RKIP, miR-29a, and miR-29b [25, 30-39]. StarBase database analysis revealed that miR-205-5p, miR-34a-5p, miR-29a, and miR-29b have no potential binding sites for circZNF532. Afterward, circZNF532 was silenced in HRMECs, resulting in only a significant decrease in PIK3CD mRNA expression (Supplementary Fig. 1). We then found a significant increase in PIK3CD mRNA expression after HG treatment and a decrease in PIK3CD mRNA expression after si-circZNF532 transfection (Fig. 2A). HG treatment led to an increase in PIK3CD protein expression and AKT phosphorylation, whereas si-circZNF532 transfection notably inhibited this effect (Fig. 2B). Actinomycin D was added to inhibit gene transcription in HRMECs. Results of qRT-PCR revealed increased mRNA stability of PIK3CD in the HG stimulation group compared with the control or osmotic group, whereas si-circZNF532 transfection repressed this increase (Fig. 2C). These findings suggested that circZNF532 may promote the mRNA stability of PIK3CD. To validate the direct interaction between circZNF532 and PIK3CD, PIK3CD-wt and PIK3CD-mut were constructed, and a luciferase reporter system was used. The result confirmed the direct binding between circZNF532 and PIK3CD, showing that circZNF532 overexpression significantly increased the luciferase activity of PIK3CD-wt but did not influence that of PIK3CD-mut (Fig. 2D). These findings indicate that circZNF532 promotes the mRNA stability of PIK3CD.

circZNF532 enhances the mRNA stability of PIK3CD. HRMECs were transfected with si-circZNF532 or si-NC and subjected to HG (25 mmol/L D-glucose) treatment for 48 h. Cells exposed to 5 mmol/L D-glucose or 25 mmol/L L-glucose served as the control or osmotic (osm) group, respectively. (A) qRT-PCR was used to detect PIK3CD mRNA; (B) Western blot analysis of PIK3CD, AKT, and p-AKT protein; (C) Actinomycin D (5 μg/mL) was added to HG-treated HRMECs with or without si-circZNF532 transfection and incubated for 0 h, 2 h, and 4 h. qRT-PCR was conducted to examine the remaining PIK3CD mRNA level. (D) The wild-type and mutant PIK3CD 3'-UTR (PIK3CD-wt and PIK3CD-mut) luciferase reporter plasmids were cotransfected into 293T cells with the circZNF532 overexpression plasmid (oe-circZNF532) or the control plasmid (oe-NC). Luciferase activity of PIK3CD was measured. Each experiment was repeated at least three times. * p < 0.05, ** p < 0.01, *** p < 0.001.
Using StarBase database analysis, circZNF532 or PIK3CD was predicted to have binding sites for TAF15 (Supplementary Fig. 2A and 2B). Next, we tested the interactions between TAF15 and circZNF532 or PIK3CD in HG-induced cells using RIP and RNA pull-down assays. The RIP data indicated a greater enrichment of circZNF532 or PIK3CD mRNA in the anti-TAF15-precipitate group than in the anti-IgG group (Fig. 3A). Consistent with this, the RNA pull-down experiment revealed preferential enrichment of TAF15 by circZNF532 sense or PIK3CD sense compared with the corresponding antisense groups (Fig. 3B). circZNF532 was overexpressed and TAF15 was silenced in HRMECs by lenti-circZNF532 infection and si-TAF15 transfection. We observed that circZNF532 overexpression elevated the PIK3CD mRNA level, which was abrogated by the knockdown of TAF15 (Fig. 3C). After treatment with actinomycin D for 2 and 4 h, the remaining PIK3CD mRNA level in each group was notably decreased. Overexpression of circZNF532 increased PIK3CD mRNA stability, whereas the knockdown of TAF15 neutralized this effect, suggesting that circZNF532 may have enhanced PIK3CD mRNA stability through TAF15 (Fig. 3D). Next, circZNF532 was silenced and TAF15 was overexpressed in HRMECs by si-circZNF532 transfection and lenti-TAF15 infection. The results demonstrated that the knockdown of circZNF532 reduced PIK3CD mRNA expression, and the simultaneous circZNF532 knockdown and TAF15 overexpression failed to change this expression (Fig. 3E). After actinomycin D treatment, circZNF532 knockdown reduced PIK3CD mRNA level and stability, but simultaneous circZNF532 knockdown and TAF15 overexpression did not affect PIK3CD mRNA expression and stability (Fig. 3F). These results indicated that circZNF532 is necessary for the regulation of PIK3CD by TAF15. Collectively, these results suggested that circZNF532 upregulated PIK3CD expression by recruiting TAF15.

circZNF532 recruits TAF15 to upregulate PIK3CD expression. HRMECs were treated with HG (25 mmol/L D-glucose) for 48 h. (A) RNA immunoprecipitation (RIP) experiments were performed using a TAF15-specific antibody or IgG. Eluted TAF15-binding RNAs (circZNF532, PIK3CD) were reverse transcribed and detected by qRT-PCR. (B) TAF15 protein pulled down with biotin-labeled circZNF532 or PIK3CD was evaluated by Western blot. (C) HRMECs were infected with lenti-circZNF532 and transfected with si-TAF15 followed by HG (25 mmol/L D-glucose) treatment for 48 h. qRT-PCR was used to measure circZNF532 expression and PIK3CD mRNA levels. (D) Actinomycin D (5 μg/mL) was added to HG-treated HRMECs with lenti-circZNF532 infection or lenti-circZNF532 infection plus si-TAF15 transfection. After incubation for 0 h, 2 h, and 4 h, qRT-PCR was conducted to detect the remaining PIK3CD mRNA levels. (E) HRMECs were transfected with si-circZNF532 and infected with lenti-TAF15 followed by HG (25 mmol/L D-glucose) treatment for 48 h. qRT-PCR was used to measure PIK3CD mRNA levels. (F) Actinomycin D (5 μg/mL) was added to HG-treated HRMECs with corresponding transfection or infection. qRT-PCR was performed to detect PIK3CD mRNA levels after incubation for 0 h, 2 h, and 4 h. The experiments were repeated at least in triplicate. * p < 0.05, ** p < 0.01, *** p < 0.001.
To further validate whether PIK3CD overexpression could rescue the biological effects of circZNF532 silencing, HRMECs were transfected with si-circZNF532 alone or with si-circZNF532 plus lenti-PIK3CD infection, followed by HG stimulation. Knockdown of circZNF532 significantly inhibited the increase in PIK3CD protein and AKT phosphorylation induced by HG, whereas the overexpression of PIK3CD restored the effect of HG treatment (Fig. 4A). In addition, circZNF532 knockdown inhibited the decline of VE-Cadherin and CD31 and restrained the increase of Vimentin and α-SMA under HG stimulation, whereas overexpression of PIK3CD recovered this HG-induced effect (Fig. 4B and 4C). Transwell and Matrigel tube formation assays showed that the depletion of circZNF532 attenuated the migratory capacity of HG-treated HRMECs and the formation of tube-like structures. However, the inhibitory effect of circZNF532 depletion on the migration and angiogenesis of HG-treated HRMECs was reversed by simultaneous circZNF532 depletion and PIK3CD overexpression (Fig. 4D and 4E). In conclusion, PIK3CD overexpression abrogated the inhibitory effect of circZNF532 silencing on EndMT, migration, and angiogenesis of HG-stimulated HRMECs.

Overexpressing PIK3CD rescues the impact of circZNF532 silencing on EndMT, migration, and angiogenesis of HG-stimulated HRMECs. HRMECs were transfected with si-circZNF532 and infected with PIK3CD followed by HG (25 mmol/L D-glucose) or 5 mmol/L D-glucose treatment (control) for 48 h. (A, B) PIK3CD, AKT, p-AKT protein, and VE-Cadherin, CD31, Vimentin, and α-SMA proteins analysis. (C) Immunofluorescent staining analysis of VE-Cadherin and α-SMA. (D) HRVEC migration was validated by Transwell assay. (E) HRVEC angiogenic ability was evaluated using the Matrigel tube formation assay. The experiments were repeated at least in triplicate. ** p < 0.01, *** p < 0.001.
DR is a leading cause of vision loss, which affects the quality of life of patients and causes a financial burden on the affected families and society at large [40]. There is increasing evidence that persistent hyperglycemia induces EndMT and abnormal endothelial cell proliferation, which may lead to retinal vascular dysfunction [41, 42]. The modulatory role of circRNAs in various human diseases, including DR, is increasingly being acknowledged. However, there is a paucity of studies exploring the mechanism by which circRNAs are involved in the regulation of EndMT in DR. In the current study, the knockdown of circZNF532 repressed HG-mediated EMT, migration, and angiogenesis in HRMECs. In terms of mechanism, we found that circZNF532 enhanced PIK3CD mRNA stability and expression through the recruitment of TAF15. Furthermore, PIK3CD overexpression rescued the inhibitory effect of circZNF532 silencing on HG-induced EndMT, migration, and angiogenesis in HRMECs.
Numerous studies have demonstrated the regulatory role of circRNAs in DR progression. In the study by Sun et al., hsa_circ_0041795 exacerbated HG-induced human retinal pigment epithelial cell (ARPE19) injury via VEGF upregulation [43]. Liu et al. reported upregulation of circRNA-cZNF609 in the high-glucose condition, and silencing of cZNF609 ameliorated vascular endothelial dysfunction and inhibited angiogenesis [44]. More importantly, Wang et al. demonstrated the upregulation of circZNF532 in patients with DR and HG-stimulated HRMECs; in addition, the knockdown of circZNF532 repressed HG-induced HRMEC proliferation, migration, and angiogenesis [22]. Liang et al. reported increased expression of circZNF532 in the serum of DR patients and the HG-treated RPE cell line ARPE-19 [45]. These findings suggest a promotive role of circZNF532 in DR progression. However, the precise impact of circZNF532 on EndMT in DR has not yet been studied. In this study, circZNF532 knockdown suppressed HG-mediated EMT progression, migration, and angiogenesis in HRMECs.
Several studies have indicated that circRNAs regulate downstream gene mRNA stability to influence the progression of diverse diseases. For example, Chen et al. reported that circRHOBTB3 restrained metastasis of colorectal cancer by regulating PTBP1 mRNA stability [46]. Pan et al. indicated that circNEIL3 facilitated glioma progression and macrophage immunosuppressive polarization by stabilizing IGF2BP3 [47]. PIK3CD is involved in regulating cell proliferation, differentiation, and biological metabolism, and its overexpression may trigger uncontrollable cell proliferation and metastasis in cancers [48, 49]. Furthermore, He et al. reported an enhanced level of PIK3CD in HG-treated HRMECs, which contributed to cell proliferation, migration, and tube formation [23]. In the present study, circZNF532 silencing inhibited PIK3CD protein expression and AKT phosphorylation in HG-stimulated HRMECs. In addition, circZNF532 silencing repressed PIK3CD mRNA stability. To the best of our knowledge, this is the first study to report this mechanism.
Subsequent bioinformatics analysis predicted that circZNF532 or PIK3CD have binding sites for TAF15. Thus, we identified the interactions in HRMECs. As expected, the enrichment of circZNF532 or PIK3CD was higher in the anti-TAF15-precipitate group than in the anti-IgG group, and TAF15 was preferentially enriched by the circZNF532 sense or PIK3CD sense groups. We next observed that circZNF532 stabilizes PIK3CD to upregulate its expression through the recruitment of TAF15. Consistent with our results, Chen et al. demonstrated that circ-ITCH recruited TAF15 protein to stabilize Nrf2 mRNA in HUVECs [50]. Finally, we found that PIK3CD overexpression abrogated the inhibitory effect of circZNF532 silencing on HG-induced cell EndMT, migration, and angiogenesis. Interestingly, in a previous study, ciRS-7 was found to upregulate PIK3CD expression to stimulate the proliferation and metastasis of oral squamous cell carcinoma [51]. Therefore, the above data suggest that circZNF532 recruited TAF15 protein to stabilize PIK3CD mRNA, thus facilitating EndMT in DR progression.
Some limitations of our study need to be considered when interpreting our findings. We have not verified our data in animal models, which requires further investigation. Currently, we are unable to obtain human diabetic retina samples to validate our experimental data in the clinical setting, which needs to be explored in future studies.
In summary, we identified a novel circRNA (circZNF532) that may affect EndMT in DR. Our findings suggest that circZNF532 recruits TAF15 to stabilize PIK3CD, which in turn promotes EndMT during DR (Supplementary Fig. 3). Our work presents a novel regulatory mechanism for DR, providing new insights for developing therapeutic approaches.
DR, Diabetic retinopathy; EndMT, Endothelial-to-mesenchymal transition; circRNAs, circular RNAs; circZNF532, circRNA zinc finger protein 532; PIK3CD, phosphoinositide-3 kinase catalytic subunit δ; TAF15, TATA box binding protein (TBP)-associated factor 15; HRMECs, human retinal microvascular endothelial cells; HG, high glucose; 3' UTR, 3' untranslated region; FBS, fetal bovine serum; EBM-2, endothelial basal medium-2; DAPI, 4',6-diamidino-2-phenylindole; SDS-PAGE, sodium dodecyl-sulfate polyacrylamide gel electrophoresis; PVDF, polyvinylidene fluoride; qRT-PCR, quantitative real-time PCR; RIP, RNA immunoprecipitation; ECL, enhanced chemiluminescence
The authors declare that they have no conflict of interest.

qRT-PCR was performed to detect mRNA levels of METTL3, SPRED2, Nrf2, AGK, PKC, PIK3CD, Sirt1, and RKIP in HRMECs after transfection with si-circZNF532 or si-NC. ** p < 0.01.
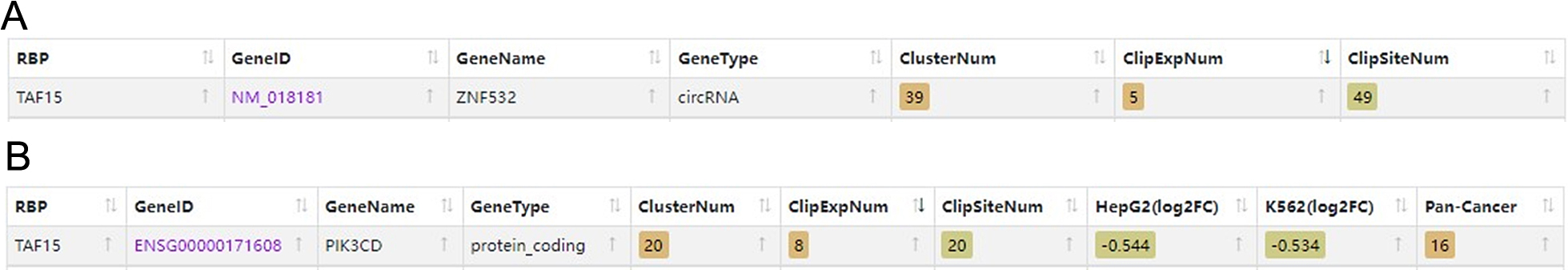
The binding relationship between TAF15 and circZNF532 (A) or PIK3CD mRNA (B) was predicted using StarBase database analysis.

A schematic illustration of the regulatory mechanism of circZNF532 in facilitating EndMT in DR. circZNF532 enhanced PIK3CD mRNA stability to upregulate its expression in HRMECs via recruitment of TAF15, and eventually facilitated EndMT in DR.