2017 年 86 巻 1 号 p. 78-86
2017 年 86 巻 1 号 p. 78-86
Transport of water into cells is mediated by plasma membrane intrinsic protein (PIP) families of aquaporin, which are involved in petal cell expansion during flower opening. In this study, we performed comprehensive characterization of aquaporin family genes and analyzed the expression of PIP genes in petals of opening flowers to examine the role of PIPs in flower opening in the carnation (Dianthus caryophyllus L.). A database search of the genome sequence revealed the existence of 26 aquaporin genes with 8 members of the PIP subfamily in the carnation ‘Francesco’. The expression of all the PIP genes was validated by the existence of expressed sequence tags, and expression analysis by quantitative reverse transcription-PCR showed that DcPIP2;1 and DcPIP1;1 are the two major PIP isoforms expressed in petals of the ‘Pure Red’ carnation. The transcripts of these two genes were also detected abundantly in other floral tissues including the calyx, style, receptacle, and ovary, as well as stems and leaves. The expression of DcPIP2;1 and DcPIP1;1 in petals was maintained at a high level throughout the flower opening process. These data suggest a putative role of these PIPs in petal growth for flower opening.
The vase life of cut ornamental flowers is the period from flower opening to senescence. In order to prolong the display time of flowers, it is an effective option to slow down the progression of flower opening, as well as to suppress senescence. In addition, some ornamentals are in high demand on specific occasions, such as carnations on Mothers’ Day. In order to develop techniques for fine control of flower opening, we need to understand the flower opening process at the molecular level, which includes the identification of the enzymes and proteins functioning in the process. For this purpose, we have been studying the molecular mechanism of flower opening using cut carnations (Dianthus caryophyllus L.) flowers as a model ornamental (Harada et al., 2010, 2011; Morita et al., 2011). We cloned and characterized several genes expressed during flower opening (Harada et al., 2010, 2011; Morita et al., 2011). Those genes encode enzymes that are involved in petal cell expansion such as xyloglucan endotransglucosylase/hydrolase (XTH), expansin, and sucrose synthase. In addition, xyloglucan oligosaccharides (XGO) treatment was found to promote flower opening in some carnation cultivars including ‘Pure Red’ in the course of our study on chemical control of flower opening in carnations (Satoh et al., 2013).
Flower opening involves petal growth, which mainly results from the expansion of petal cells (Kenis et al., 1985; Koning, 1984; Norikoshi et al., 2013). Previous studies demonstrated sugar accumulation (Evans and Reid, 1988; Ho and Nichols, 1977; Norikoshi et al., 2013; Yamada et al., 2009) and a decrease in osmotic potential in petals during flower opening (Norikoshi et al., 2013; Yamada et al., 2009), which is thought to contribute to cell expansion by promoting water influx into the petal cells. Influx of water into cells is mediated by channel protein aquaporins (AQPs) (Maurel et al., 2008). These are small membrane proteins involved in transport of water and low molecular weight solutes across biological membranes. Plant AQPs are encoded by a large family (35 AQPs in Arabidopsis, 31 in maize, and 33 in rice; Chaumont et al., 2000; Johanson et al., 2001; Sakurai et al., 2005), and divided into 5 subfamilies; the plasma membrane intrinsic proteins (PIPs), the tonoplast intrinsic proteins (TIPs), the nodulin-26-like intrinsic membrane proteins (NIPs), the small basic intrinsic proteins (SIPs), and the X-intrinsic proteins (XIPs) (Danielson and Johanson, 2008; Maurel et al., 2008). Although XIPs are absent in Arabidopsis, maize, and rice, they are found in a moss, Physcomitrella patens (Danielson and Johanson, 2008), some Solanaceae species including tobacco and tomatoes (Bienert et al., 2011; Reuscher et al., 2013), and soybeans (Zhang et al., 2013). Water transport across the plasma membrane is mediated by PIPs, which are divided into PIP1 and PIP2 subgroups according to their differences in primary structure. PIP1 and PIP2 have different functional properties when expressed in Xenopus oocytes; i.e. PIP2 shows high water transport activity while PIP1s show low or no water permeability (Chaumont et al., 2000; Daniels et al., 1994; Fetter et al., 2004; Yamada et al., 1995). However, PIP1 shows water permeability by interacting with PIP2 (Fetter et al., 2004), particularly by forming a heterotetramer with PIP2 (Yaneff et al., 2014). Both PIP1 and PIP2 have important roles in water transport across the plasma membrane.
Water transport by PIPs is associated with tissue expansion and elongation in plants. PIP was found to be involved in petiole elongation during leaf unfolding in tobacco by antisense suppression of the NtAQP1 gene (Siefritz et al., 2004). In tulips, temperature-dependent flower opening and closing is associated with water uptake by petal tissues, which is suggested to involve AQP activity (Azad et al., 2004). Also direct evidence of the participation of a PIP (RhPIP2;1) in petal cell expansion was demonstrated in rose flowers (Ma et al., 2008). It is reasonable to anticipate the presence of genes for PIP that participate in water uptake during petal growth of opening carnation flowers. This notion is supported by our previous finding that the expression of a PIP isoform was induced during flower opening in carnation petals (Harada et al., 2010). However, we do not know yet how many PIP genes exist in carnations and which of those participate in water uptake during petal growth of opening carnation flowers, since carnation AQP genes have not yet been studied extensively.
The carnation genome sequence was recently determined in the cultivar ‘Francesco’ (Yagi et al., 2014), which enabled us to perform comprehensive analysis of the carnation AQP family. In the present study, we aimed to characterize AQP genes expressed in carnation flowers in silico using the carnation genome sequence in order to better understand the flower opening process at the molecular level. We analyzed the expression of carnation PIP genes (DcPIPs) to identify PIP isoforms expressed abundantly in petals of opening flowers. In addition, we examined the expression of PIP genes in XGO-treated flowers on the assumption that the increase in water uptake in petal cells may be associated with the enhancement of flower opening by XGO treatment.
Cut carnation flowers (D. caryophyllus L. ‘Pure Red’) that belong to the spray category of carnation flower were harvested when the first flower out of 6 to 7 flower buds was nearly open at the nursery of a commercial grower in Miyagi prefecture, Japan. The flowers were transported dry to the laboratory at the Biotechnology Research Department, Kyoto Prefectural Agriculture, Forestry and Fisheries Technology Center in Kyoto prefecture on the day after harvest. They were put in plastic containers with their cut end in tap water under continuous light from white fluorescent lamps (14 μmol·m−2·s−1) at 23°C.
The flower opening process was separated into 6 stages according to Harada et al. (2010): opening stage 1 (Os 1), petals just emerged from buds; Os 2, petals elongated vertically; Os 3, petal clusters expanded; Os 4, outer petals start to be reflexed (bend outwards); Os 5, outer petals have been reflexed; Os 6, fully open flower with outer petals at right angles to a stem. Ten outermost petals per flower were sampled at respective stages, and stored at −80°C for extraction of RNA. Other tissues, including leaves, stems, calyx, ovary, and style were sampled from plants with fully opened flowers and stored as above. Stamens are absent from ‘Pure Red’ carnation flowers. In carnation flowers, the stigma is along the surface of the style (rather than a distinct structure like in other flowers). Therefore, the term ‘style’ was used to describe the style plus stigma in this study. Three independent samples of each tissue were collected at each stage.
Carnation flowers at Os 2 were used for XGO treatment. Samples of 6 flowers with 15 cm-long stems were placed with their cut stem ends in a 50-mL plastic tube with 30 mL test solution. The control solution was distilled water, and the test solution contained the XGO mixture at 1% in distilled water. The XGO mixture consisted of XG7 (xyloglucan heptaoligosaccharide), XG8 (xyloglucan octaoligosaccharide), and XG9 (xyloglucan nonaoligosaccharide) at a ratio of 1:4:5 was prepared from tamarind seed gum as described previously (Satoh et al., 2013). The flowers were left under continuous light from white fluorescent lamps (14 μmol·m−2·s−1) at 23°C. After two days, flower opening was scored by determining the flower opening stage for each flower in the sample. Petals were sampled from the flowers and stored at −80°C as described above. In this experiment, 6 flowers were treated with XGO, and petals taken from 2 flowers were mixed to make one sample so that three independent samples were collected.
Database search for carnation AQPsThe amino acid sequences of 35 AQPs from Arabidopsis (Johanson et al., 2001) listed in The Arabidopsis Information Resource (TAIR10; Lamesch et al., 2012; http://www.arabidopsis.org) were retrieved through the website of the Ensembl Genomes database (Release 30; Kersey et al., 2016; http://ensemblgenomes.org/). The amino acid sequences of XIPs from tobacco (XIP1;1α and XIP1;1β; Bienert et al., 2011), soybeans (GmXIP1;1 and GmXIP1;2; Zhang et al., 2013), and tomatoes (SlXIP1;1-SlXIP1;6; Reuscher et al., 2013) were also retrieved from the Ensembl Genomes. These sequences were used as queries for a BLAST (Altschul et al., 1997) search to identify open reading frames (ORFs) encoding AQP in the carnation genome sequence database (Carnation DB; Yagi et al., 2014; http://carnation.kazusa.or.jp/index.html). ORFs with an E-value less than 1e-30 were retrieved as candidates for carnation AQPs. In order to exclude redundant sequences, highly similar ORFs (similarity threshold: 98%) were clustered using the CD-HIT program (Li and Godzik, 2006). Sequence similarity of the clustered ORFs was confirmed by comparison of the genomic sequences (scaffold sequences) of each ORF using GENETYX software (GENETYX, Tokyo, Japan). Scaffold sequences of the carnation DB were also searched by the TBLASTN program using the carnation AQP ORFs as queries. Short ORFs and a scaffold encoding polypeptides that lack Asn-Pro-Ala (NPA) motifs, which are highly conserved in AQPs and located in the pore of the channels (Maurel et al., 2008), were excluded since they were not supposed to be functional AQPs.
Carnations expressed sequence tags (ESTs) homologous to each AQP gene were searched for by BLASTN in the NCBI sequence database (http://blast.ncbi.nlm.nih.gov). Considering possible sequence variations in different alleles and cultivars, EST clones with an identity higher than 98% were regarded as cDNAs derived from an identical AQP gene. To estimate the expression level of each gene, the numbers of the ESTs from leaves and flowers were counted.
Multiple alignment of carnation AQPs was performed by ClustalW (Larkin et al., 2007) on the website of the DDBJ (http://clustalw.ddbj.nig.ac.jp) by the neighbor-joining method with a default setting (ver 2.1), and a phylogenic tree was constructed using TreeView X software.
Cloning of cDNAs encoding PIP homologsTotal RNA was extracted from 1 to 2 g of frozen petals according to Harada et al. (2005). Single strand cDNA was synthesized from the total RNA with ReverTra Ace reverse transcriptase (Toyobo, Osaka, Japan) using an oligo dT primer. For amplification of full length cDNAs of DcPIP2;1, a primer set (see Table 1) was designed in the 5'- and 3'-untranslated regions according to the sequence of full length cDNA of PIP2 from ‘Master’ carnation (accession no. GU989036). A primer set for full length DcPIP1;1 cDNA was also designed based on the EST sequences (accession nos. FY403342 and FY394588) corresponding to the DcPIP1;1 ORF (Dca52692.1; listed in the Carnation DB). Full-length cDNAs were amplified by polymerase chain reaction (PCR) using a high-fidelity DNA polymerase, KOD plus (Toyobo). The amplified fragments were cloned into the pGEM-T vector (Promega, Madison, WI, USA) after A-tailing with Taq polymerase (Toyobo). The entire sequence of the cDNA was confirmed by sequencing on both strands.
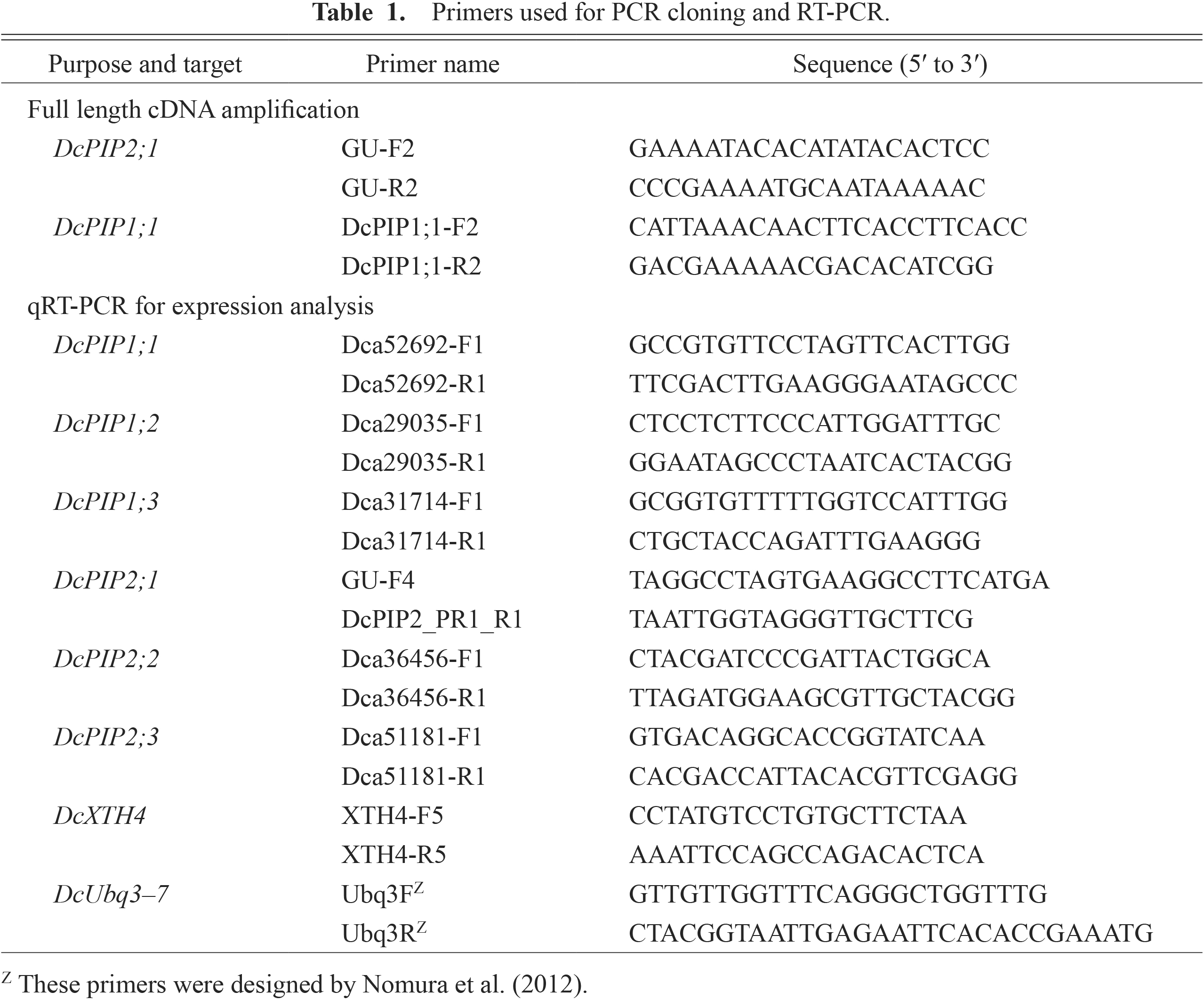
Primers used for PCR cloning and RT-PCR.
Total RNA was extracted from frozen samples and cDNA synthesis was performed as described above. To analyze the transcript level of DcPIP and DcXTH4 genes, primer sets (listed in Table 1) were designed in the 3'-end region of the ORF of each gene. Partial cDNA fragments of respective genes were amplified from petal cDNAs by PCR using the primer sets. After confirming the sequences of the fragments by direct sequencing, they were cloned into the pGEM-T vector (Promega). The resulting plasmids were used as a standard for quantification. Quantitative reverse transcription-PCR (qRT-PCR) was performed with a DNA Engine OPTICON 2 real-time PCR cycler (MJ Research, Waltham, MA, USA) using the LightCycler FastStart DNA Master PLUS SYBR Green I kit (Roche, Basel, Switzerland) or KOD SYBR qPCR Mix (Toyobo) following the manufacturer’s instructions. The transcript level was calculated using a dilution series of standard plasmids. The transcript level of ubiquitin genes (DcUbq3 to DcUbq7; Nomura et al., 2012) was also measured and used for normalization of the PIP transcript level. Analyses were conducted with three independent replicates for each sample, and data were given by the mean ± SD.
The carnation genome sequence was determined in the cultivar ‘Francesco’ (Yagi et al., 2014). We first characterized AQP genes encoded in the ‘Francesco’ genome by an in silico search. For this purpose, a BLAST search was performed for carnation ORFs in the Carnation DB using amino acid sequences of Arabidopsis AQPs and XIPs from tobacco, soybeans, and tomatoes as queries. After excluding redundant sequences by clustering and incomplete ORFs that lack NPA motifs, we obtained 26 ORFs as AQP homologs in the carnation (Table 2). During the clustering process, 8 ORFs with highly similar sequences were classified into 4 clusters. We retrieved the genomic sequences (coding region plus 2 kb-upstream and 2 kb-downstream regions) of these ORFs from the Carnation DB and compared them. We found that the sequences in the same clusters were highly homologous throughout the coding and flanking regions (sequence identities were 81–99%). Therefore, we considered each member of the cluster to be putative alleles of identical loci (Table 2). We also performed a BLAST search for scaffold sequences in the Carnation DB using these ORFs as queries. However, a short sequence that encoded only 148 amino acids, in which the 1st NPA motif was missing, was found. Therefore, we finally identified 26 AQPs in the carnation. Multiple alignment revealed that they were classified into 5 subfamilies including 8 PIPs, 11 TIPs, 4 NIPs, 2 SIPs, and a XIP (Fig. 1). The PIP subfamily contained 3 and 5 members of the PIP1 and PIP2 subgroups, respectively (Fig. 1), and they were designated according to their expression level revealed by the following expression analysis. The nomenclature of the members of the TIP and SIP subfamilies was based on the homology with Arabidopsis AQPs. NIPs were classified into subgroups by phylogenetic analysis with NIPs from Arabidopsis, soybeans, and tomatoes (data not shown). It is noted that 4 members of the carnation NIPs were classified as NIP5 and NIP6, and there were no members for NIP1-NIP4.

Results of a database search for carnation AQPs.

Phylogenetic analysis of AQPs in carnations. The amino acid sequences of 26 AQPs identified in the Carnation DB were aligned using ClustalW by the neighbor-joining method and the phylogenetic tree was constructed by TreeView X. The scale indicates the base-substitution rate.
In order to confirm the expression of the carnation AQP genes and estimate their expression levels, we searched for ESTs corresponding to each AQP gene in the sequence database. As a result, ESTs for 15 AQP genes were found (Table 2). Most of the ESTs found were derived from the work by Yagi et al. (2012) and isolated from flower buds and leaves of ‘Francesco’. The number of ESTs suggest that DcPIP1;1, DcPIP2;1, and DcTIP2;2 are expressed abundantly in flowers. The members of the TIP subfamily function in transport of water as well as ammonia, hydrogen peroxide, and urea (Hove and Bhave, 2011; Sade et al., 2009). The motifs related to substrate selectivity of AQPs, ar/R motifs (Wallace and Roberts, 2004) and P1-P5 residues (Froger et al., 1998) were searched for in the sequence of DcTIP2;2. The sequences were H-I-G-R for the ar/R motif and T-S-A-Y-W for P1-P5 residues. These were identical to the residues of AtTIP2;1 and AtTIP2;3 that have transport activities of ammonia and hydrogen peroxide (Hove and Bhave, 2011), suggesting that DcTIP2;2 may be involved in transport of ammonia or hydrogen peroxide. Since water uptake into cells across the plasma membrane is mediated by PIPs, we further focused on PIP isoforms expressed in carnation flowers.
The expression of all the DcPIP genes was confirmed by the existence of ESTs, but their expression levels were suggested to be different since the number of ESTs found was variable from 1 to 81 for each gene. Therefore, we determined the expression level of the DcPIP genes in petals and leaves by qRT-PCR. Excluding DcPIP2;4 and DcPIP2;5 with putatively low expression levels estimated by the number of ESTs (1 and 3 ESTs, respectively), the expression of PIP genes was analyzed using petals of opening flowers and green leaves sampled from cut flowers. We used a cultivar ‘Pure Red’ in this study since the enhancement of flower opening by XGO treatment was prominent in this cultivar in our previous study (Satoh et al., 2013). As a result, DcPIP2;1 and DcPIP1;1 showed high expression levels in both petals and leaves compared with other DcPIPs in ‘Pure Red’ (Fig. 2). DcPIP1;3 corresponds to a carnation PIP gene (accession no. AB517656) identified as a differentially expressed gene during the flower opening process by suppression subtractive hybridization, and its expression is induced from Os 3 to Os 6 (Harada et al., 2010). However, the expression level of DcPIP1;3 is lower than that of DcPIP1;1. These results were consistent with the EST search results and indicate that DcPIP2;1 and DcPIP1;1 are two major PIP isoforms expressed in carnations. It is likely that these PIP isoforms are involved in the uptake of water into petal cells, since both PIP2 and PIP1 have water transport activity in heterotetramers (Yaneff et al., 2014).

Expression of DcPIP genes in petals and leaves in the ‘Pure Red’ carnation. Total RNA was prepared from petals and leaves, and qRT-PCR was performed to quantify the transcript level of DcPIP genes. For petal samples, an equal amount of cDNAs from petals at Os 1 to Os 6 was mixed and used for qPCR. The transcript level of DcPIPs was normalized with that of DcUbq3–7. Data shown are mean ± SD (n = 3).
Next, we determined the sequences of DcPIP2;1 and DcPIP1;1 in ‘Pure Red’ by cloning full length cDNAs. The nucleotide sequence of DcPIP1;1 cDNA from ‘Pure Red’ was completely identical to that of ORF listed in the Carnation DB (Dca52692.1) and ESTs from ‘Francesco’. As for PIP2;1, there is a full length cDNA from ‘Master’ carnation (DcPIP2;1-Master; accession no. GU989036) in the sequence database. We compared the nucleotide sequences of the DcPIP2;1 cDNA cloned from ‘Pure Red’ (DcPIP2;1-PR) and that of DcPIP2;1-Master, and found that there were 2 indels and 10 nucleotide substitutions in a 983 bp sequence. These differences resulted in an indel and an amino acid sequence substitution (data not shown), indicating that there are some variations in PIP2;1 sequences among cultivars. Two NPA motifs that are highly conserved in AQPs and located in the pore of the channels (Maurel et al., 2008) were conserved in both DcPIP1;1 and DcPIP2;1 (data not shown). The nucleotide sequences of DcPIP2;1 and DcPIP1;1 cDNA from ‘Pure Red’ were deposited in DDBJ, EMBL, and NCBI sequence databases under accession nos. LC062382 (DcPIP1;1) and LC062383 (DcPIP2;1-PR).
Expression profiles of DcPIP2;1 and DcPIP1;1 in carnation flowersIn order to elucidate the expression profiles of DcPIP2;1 and DcPIP1;1 in tissues, we performed qRT-PCR analysis of these genes in various tissues of carnation flowers (calyx, style, receptacle, and ovary) as well as stems and leaves. Both DcPIP2;1 and DcPIP1;1 were expressed not only in petals, but also in other tissues (Fig. 3). A high expression level of DcPIP2;1 was observed in all the tissues examined, suggesting that DcPIP2;1 is the primary PIP isoform expressed in the aerial part of carnation plants. However, it is noted that the expression level of DcPIP1;1 was highest in petals than in any other tissues, which suggests that DcPIP1;1 plays a role in water transport in petals.

Expression of DcPIP2;1 and DcPIP1;1 in various tissues of the ‘Pure Red’ carnation at Os 6 (full-open stage). Total RNA was prepared from the tissues indicated and qRT-PCR was performed as in Figure 2. Data shown are mean ± SD (n = 3). The different letters indicate significant differences (Tukey’s test: P < 0.05).
Next, we examined changes in expression levels of DcPIP2;1 and DcPIP1;1 during flower opening. The transcript level of DcPIP2;1 did not show significant changes during the flower opening process (Fig. 4A). Also, the expression of DcPIP1;1 tended to increase at Os 3 although there was no statistical difference throughout flower opening (Fig. 4B). The expression of DcPIP1;3 was shown to be elevated at Os 5 and Os 6 (Harada et al., 2010). Considering that DcPIP2;1 and DcPIP1;1 are expressed more abundantly than DcPIP1;3 as shown in Figure 2, the stable expression of DcPIP2;1 and DcPIP1;1 suggests that the expression level of these PIPs is high enough for flower opening. In rose petals, the role of PIP2 (Rh-PIP2;1) in the regulation of petal cell expansion was demonstrated (Ma et al., 2008). Since the expression of DcPIP2;1 is highest among DcPIP isoforms, it is plausible that DcPIP2;1 participates in petal growth in carnation flowers like Rh-PIP2;1 in rose petals. In addition, water transport activity of PIP2 is regulated by post-translational mechanisms such as phosphorylation, cellular trafficking, and heterotetramer formation (Maurel et al., 2008), and phosphorylation of plasma membrane-localized AQP is thought to be involved in the regulation of tulip flower opening and closing (Azad et al., 2004). Therefore, it is also plausible that the activity of DcPIP2;1 is regulated by a post-trasnlational mechanism during flower opening.
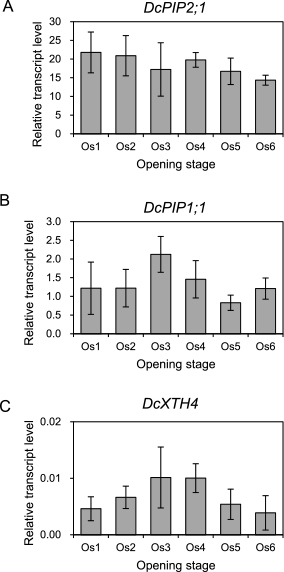
Expression of DcPIP2;1 (A), DcPIP1;1 (B), and DcXTH4 (C) during flower opening in the ‘Pure Red’ carnation. Total RNA was prepared from petals at each stage and qRT-PCR was performed as in Figure 2. Data shown are mean ± SD (n = 3). No significant difference was observed among the transcript levels at each stage (Tukey’s test: P < 0.05).
In a previous study, we observed that XGO treatment enhanced flower opening in the ‘Pure Red’ carnation (Satoh et al., 2013). On the assumption that water uptake in petal cells may be associated with the enhancement of flower opening by XGO, we examined the expression of DcPIP2;1 and DcPIP1;1 in XGO-treated flowers. In this study, flowers at Os 2 were treated with 1% XGO for 2 days. Significant enhancement of flower opening was observed by XGO treatment as in the previous study, with the median flower-opening score of control flowers being 3.5 and that of XGO-treated flowers being 6 (Fig. 5A).

Gene expression analysis in XGO-treated flowers of the ‘Pure Red’ carnation. (A) Flowers at Os 2 were treated with (+XGO) or without (−XGO) 1% XGO solution for 2 days. Flower-opening score (the flower opening stage for each flower) at the end of the treatment is shown by a box plot (n = 6). The values of the maximum, third quartile, and median were all 6 for XGO-treated flowers. An asterisk indicates significant difference between non-treated and treated samples by Mann-Whitney’s U-test at P < 0.05. Expression of DcPIP2;1 (B), DcPIP1;1 (C), and DcXTH4 (D) in petals of flowers treated with or without XGO for 2 d. “0 d” indicates flowers sampled at the onset of the treatment. Data shown are mean ± SD (n = 3). No significant difference was observed among the transcript levels of each sample for (B)–(D) (Tukey’s test: P < 0.05).
Using petals sampled from these flowers, expression of DcPIP2;1 and DcPIP1;1 was examined by qRT-PCR. There were no significant differences in the transcript level of these genes among the initial samples before treatment (0 d-sample), and XGO-treated and untreated samples (Fig. 5B, C). It is reasonable that there was no difference in DcPIP2;1 expression since it was unchanged during flower opening (Fig. 4A). Although expression of DcPIP1;1 increased at Os 3 (Fig. 4B), its expression level was similar in the 0 d-sample and untreated sample. This may be due to the variation in flower opening stage of the untreated samples with flower-opening scores from 2 to 6 (Fig. 5A). This suggests that the changes in the transcript level caused by XGO treatment are not larger than those caused by the variation in flower opening stage. It is also possible that changes in gene expression due to XGO treatment occur in shorter period than 2 days and that we may have failed to detect the early response. In any case, our data suggest that XGO treatment has little influence on DcPIP2;1 and DcPIP1;1 expression.
In order to investigate whether XGO treatment accelerates the flower opening process by modulating the expression of the genes involved in petal cell expansion, we examined the expression of a XTH gene in XGO-treated flowers. There are 4 XTH genes (DcXTH1 to DcXTH4) expressed in petals of opening carnation flowers (Harada et al., 2011). We selected DcXTH4 for this analysis since its expression steadily increased during flower opening with the highest expression at Os 6 in ‘Light Pink Barbara’ (Harada et al., 2011). The expression of DcXTH4 during flower opening in ‘Pure Red’ was examined, and the result revealed that its transcript level tended to increase at Os 3 and Os 4 (Fig. 4C). Although there was a discrepancy in the expression pattern between ‘Light Pink Barbara’ and ‘Pure Red’, the expression level of DcXTH4 was different between Os 3–4 and Os 5–6 in ‘Pure Red’. We assumed that this gene could be a marker for petal expansion, which is prominent at Os 3 to 4. We then examined the expression of DcXTH4 in petals of flowers treated with XGO for 2 days. There were no differences in the transcript level of DcXTH4 among the 0 d-, untreated, and treated samples as well as DcPIP2;1 and DcPIP1;1 (Fig. 5D). Our data thus suggest that XGO treatment does not enhance the expression of genes associated with flower opening. XGO may affect flower opening by modulation of cell wall expansion through incorporation of XGO into cell walls by xyloglucan endotransglycosylase activity of XTH, as proposed by Satoh et al. (2013), rather than up-regulation of the genes involved in petal growth.
In summary, we identified a carnation AQP family by an in silico search and characterized PIP genes expressed in flowers of the ‘Pure Red’ carnation. Our data revealed that DcPIP2;1 and DcPIP1;1 are the two major isoforms in petals of opening flowers and that their expression levels were maintained at a high level throughout flower opening, suggesting a putative role of these PIPs in petal growth for flower opening.